3OOK
 
 | |
3OKI
 
 | |
3OMM
 
 | |
3OKH
 
 | |
3OMK
 
 | |
9GOX
 
 | | Crystal structure of Fab B6-D9 in complex with CD38 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Dreyfus, C, Freier, R. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biparatopic binding of ISB 1442 to CD38 in trans enables increased cell antibody density and increased avidity.
Mabs, 17, 2025
|
|
9GOY
 
 | | Crystal structure of Fab E2-RecA in complex with CD38 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, CHLORIDE ION, Fab E2-RecA heavy chain, ... | | Authors: | Dreyfus, C, Fritz, G. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biparatopic binding of ISB 1442 to CD38 in trans enables increased cell antibody density and increased avidity.
Mabs, 17, 2025
|
|
3OLF
 
 | |
6GE6
 
 | |
6GE4
 
 | |
6GE5
 
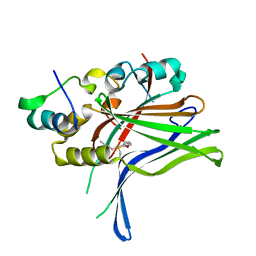 | |
6GEC
 
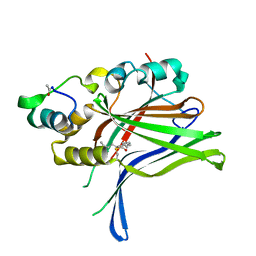 | |
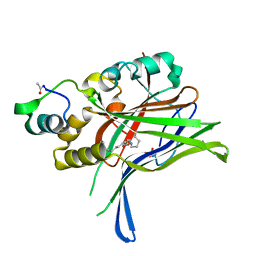
6GEI
 
 | |
6GEG
 
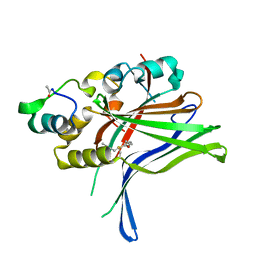 | |
6GEK
 
 | |
6GEE
 
 | |
6GE3
 
 | |
1OP8
 
 | | Crystal Structure of Human Granzyme A | | Descriptor: | Granzyme A, SULFATE ION | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Bode, W, Jenne, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the apoptosis-inducing human granzyme A dimer
NAT.STRUCT.BIOL., 10, 2003
|
|
5OAQ
 
 | |
