5YU7
 
 | |
5YU6
 
 | | CRYSTAL STRUCTURE OF EXPORTIN-5:RANGTP COMPLEX | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Yamazawa, R, Jiko, C, Lee, S.J, Yamashita, E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Basis for Selective Binding of Export Cargoes by Exportin-5
Structure, 26, 2018
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5W1B
 
 | | Tryptophan indole-lyase | | Descriptor: | POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6J4P
 
 | |
6J4V
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4Q
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4O
 
 | |
6J4S
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3O0G
 
 | | Crystal Structure of Cdk5:p25 in complex with an ATP analogue | | Descriptor: | Cell division protein kinase 5, Cyclin-dependent kinase 5 activator 1, {4-amino-2-[(4-chlorophenyl)amino]-1,3-thiazol-5-yl}(3-nitrophenyl)methanone | | Authors: | Mapelli, M. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining Cdk5 ligand chemical space with small molecule inhibitors of Tau phosphorylation
Chem.Biol., 12, 2005
|
|
6J4U
 
 | |
7CAZ
 
 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CAW
 
 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-ACP reductase FabG, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CAX
 
 | | Crystal structure of bacterial reductase | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-ACP reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7F28
 
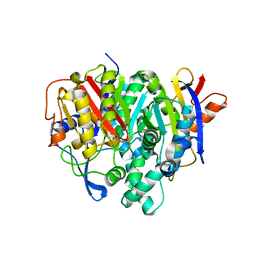 | | Crystal structure of a bacterial ketosynthase | | Descriptor: | Ketoacyl_synth_N domain-containing protein, Putative 3-oxoacyl-[ACP] synthase FabV | | Authors: | Lee, W.C, Kim, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
7F27
 
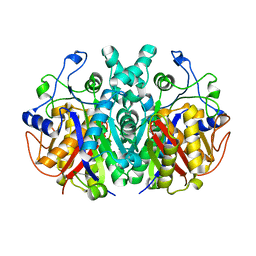 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
4NKR
 
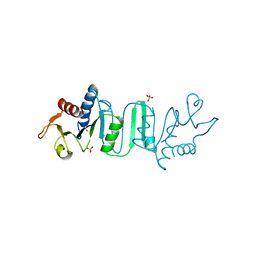 | | The Crystal structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Choe, J, Kim, D, Choi, S, Kim, H. | | Deposit date: | 2013-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insight into the Role of E.coli MobB in Molybdenum Cofactor Biosynthesis based on the high resolution crystal structure
TO BE PUBLISHED
|
|
