6WXO
 
 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 2-histidine 2-glutamate centre TFD-HE | | Descriptor: | GLYCEROL, SULFATE ION, TFD-HE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WRX
 
 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRV
 
 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | Authors: | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRW
 
 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5K7V
 
 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein HR00C3 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.165 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5KBA
 
 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein ANK1C2 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-06-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
6WVS
 
 | | Hyperstable de novo TIM barrel variant DeNovoTIM15 | | Descriptor: | DeNovoTIM15 hyperstable de novo TIM barrel | | Authors: | Bick, M.J, Haydon, I.C, Caldwell, S.J, Zeymer, C, Huang, P, Fernandez-Velasco, D.A, Baker, D. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KWD
 
 | |
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MSP
 
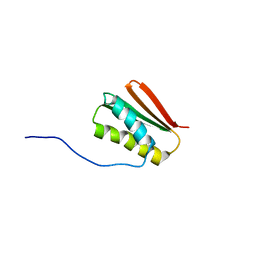 | | De novo Designed Protein Foldit3 | | Descriptor: | De novo Designed Protein Foldit3 | | Authors: | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | Deposit date: | 2018-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
6MRS
 
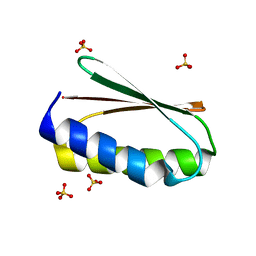 | |
6MSR
 
 | | Crystal structure of pRO-2.5 | | Descriptor: | pRO-2.5 | | Authors: | Bick, M.J, Sankaran, B, Boyken, S.E, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
1YSD
 
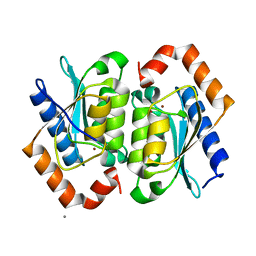 | | Yeast Cytosine Deaminase Double Mutant | | Descriptor: | CALCIUM ION, Cytosine deaminase, ZINC ION | | Authors: | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | Deposit date: | 2005-02-08 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
6NAF
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Selvaraj, B, Park, J, Cuneo, M.J, Myles, D.A.A, Baker, D. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.923 Å), X-RAY DIFFRACTION | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
1YSB
 
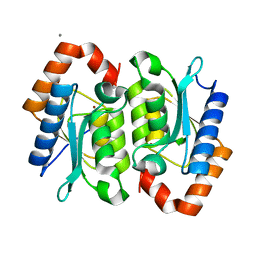 | | Yeast Cytosine Deaminase Triple Mutant | | Descriptor: | CALCIUM ION, Cytosine deaminase, ZINC ION | | Authors: | Korkegian, A, Black, M.E, Baker, D, Stoddard, B.L. | | Deposit date: | 2005-02-08 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational thermostabilization of an enzyme.
Science, 308, 2005
|
|
6X9Z
 
 | |
5BRW
 
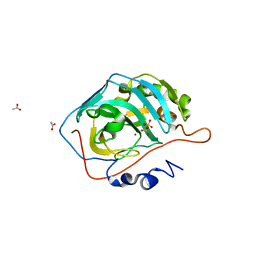 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BRU
 
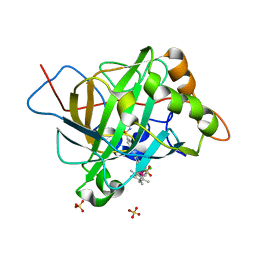 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION, ... | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
3J03
 
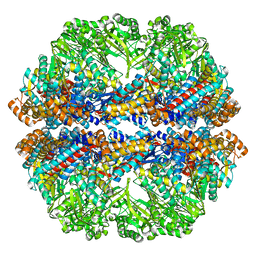 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMX
 
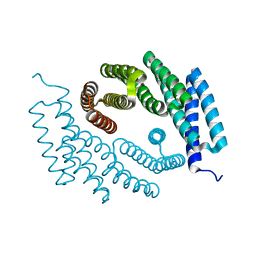 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
