5JHN
 
 | | Structure of G9a SET-domain with Histone H3K9Ala mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9A mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIN
 
 | | Structure of G9a SET-domain with Histone H3K9M mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9M mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JLE
 
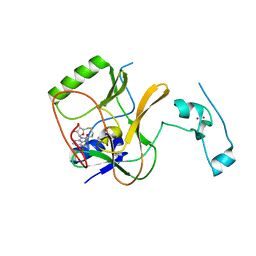 | | Crystal structure of SETD2 bound to SAH | | Descriptor: | Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, H, Yang, S, Zheng, X. | | Deposit date: | 2016-04-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
5JLB
 
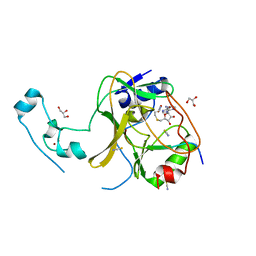 | |
5JIY
 
 | | Structure of G9a SET-domain with Histone H3K9norLeucine mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 mutant peptide with H3K9nor-leucine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JJY
 
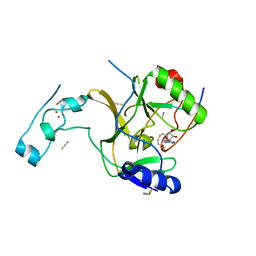 | | Crystal structure of SETD2 bound to histone H3.3 K36M peptide | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yang, S, Zheng, X, Li, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
2M0O
 
 | |
2PVC
 
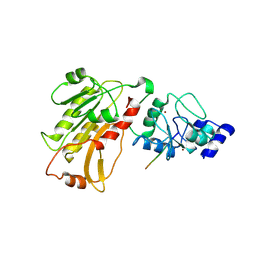 | | DNMT3L recognizes unmethylated histone H3 lysine 4 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, Histone H3 peptide, ZINC ION | | Authors: | Cheng, X. | | Deposit date: | 2007-05-09 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
2PV0
 
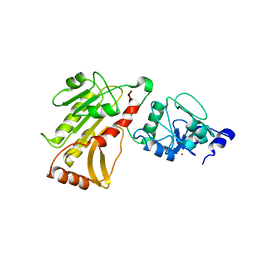 | | DNA methyltransferase 3 like protein (DNMT3L) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, ZINC ION | | Authors: | Cheng, X. | | Deposit date: | 2007-05-09 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
3QL9
 
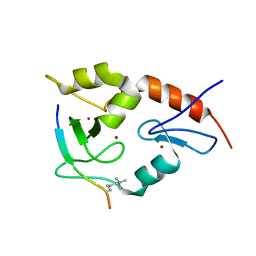 | |
3QLN
 
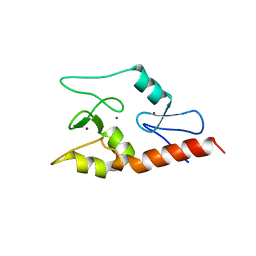 | | Crystal structure of ATRX ADD domain in free state | | Descriptor: | Transcriptional regulator ATRX, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QLA
 
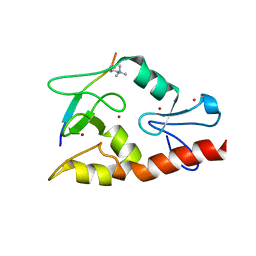 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QZS
 
 | |
3QZV
 
 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | Descriptor: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QLC
 
 | |
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
2KGG
 
 | |
2KGI
 
 | |
2KU7
 
 | |
