5AZ2
 
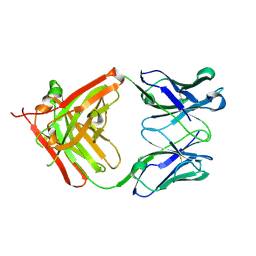 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5AVH
 
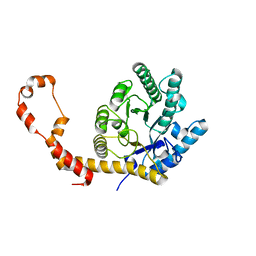 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVN
 
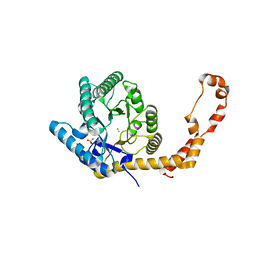 | | The 1.03 angstrom structure (P212121) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5H5G
 
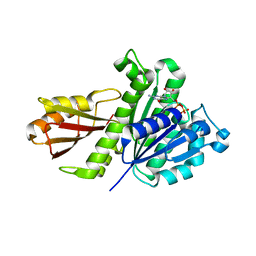 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5H5I
 
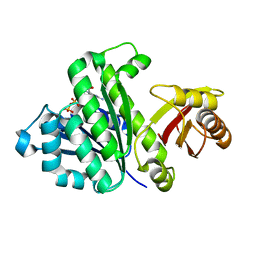 | | Staphylococcus aureus FtsZ-GDP R29A mutant in R state | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5AVD
 
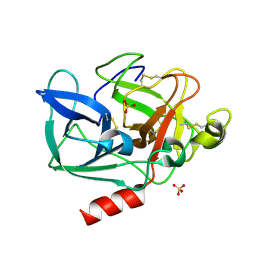 | | The 0.86 angstrom structure of elastase crystallized in high-strength agarose hydrogel | | Descriptor: | Chymotrypsin-like elastase family member 1, SULFATE ION | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVG
 
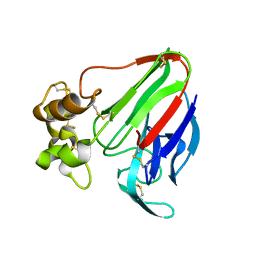 | | The 0.95 angstrom structure of thaumatin crystallized in high-strength agarose hydrogel | | Descriptor: | Thaumatin-1 | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Hirose, M, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5H5H
 
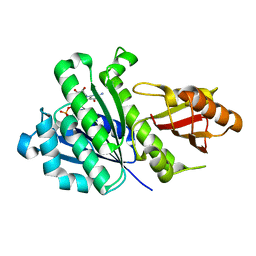 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
8HTB
 
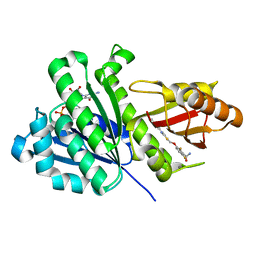 | | Staphylococcus aureus FtsZ 12-316 complexed with TXH9179 | | Descriptor: | 3-[(6-ethynyl-[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Bryan, E, Ferrer-Gonzalez, E, Sagong, H.Y, Fujita, J, Mark, L, Kaul, M, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Antibacterial Characterization of a New Benzamide FtsZ Inhibitor with Superior Bactericidal Activity and In Vivo Efficacy Against Multidrug-Resistant Staphylococcus aureus.
Acs Chem.Biol., 18, 2023
|
|
7XTX
 
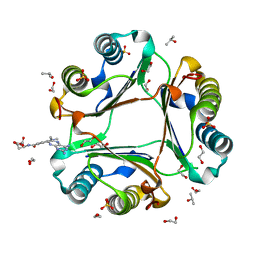 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
7XVX
 
 | |
1FIY
 
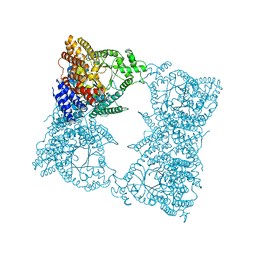 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3VGW
 
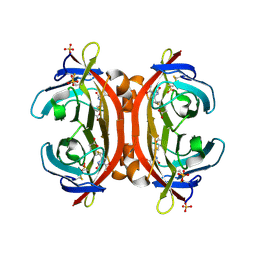 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VHH
 
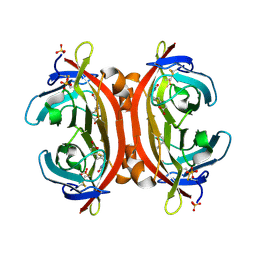 | | Crystal structure of DiMe-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1,3-dimethyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
7EV5
 
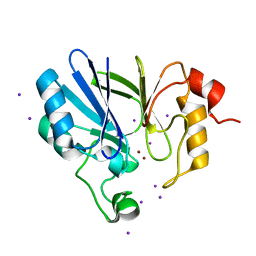 | | Crystal structure of BLEG-1 B3 metallo-beta-lactamase | | Descriptor: | IODIDE ION, Lactamase_B domain-containing protein, ZINC ION | | Authors: | Au, S.X, Muhd Noor, N.D, Matsumura, H, Rahman, R.N.Z.R.A, Normi, Y.M. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Dual Activity BLEG-1 from Bacillus lehensis G1 Revealed Structural Resemblance to B3 Metallo-beta-Lactamase and Glyoxalase II: An Insight into Its Enzyme Promiscuity and Evolutionary Divergence.
Int J Mol Sci, 22, 2021
|
|
1V40
 
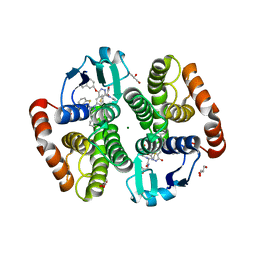 | | First Inhibitor Complex Structure of Human Hematopoietic Prostaglandin D Synthase | | Descriptor: | 3-(1,3-BENZOTHIAZOL-2-YL)-2-(1,4-DIOXO-1,2,3,4-TETRAHYDROPHTHALAZIN-6-YL)-5-[(E)-2-PHENYLVINYL]-3H-TETRAAZOL-2-IUM, GLUTATHIONE, GLYCEROL, ... | | Authors: | Inoue, T, Okano, Y, Kado, Y, Aritake, K, Irikura, D, Uodome, N, Kinugasa, S, Okazaki, N, Matsumura, H, Kai, Y, Urade, Y. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First determination of the inhibitor complex structure of human hematopoietic prostaglandin D synthase.
J.Biochem.(Tokyo), 135, 2004
|
|
6K76
 
 | |
6K78
 
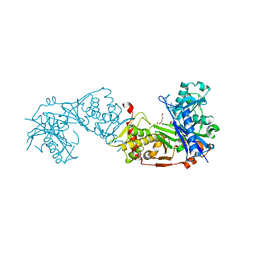 | | Glycerol kinase form Thermococcus kodakarensis, complex structure with substrate. | | Descriptor: | GLYCEROL, Glycerol kinase, TRIETHYLENE GLYCOL | | Authors: | Koga, Y, Angkawidjaja, C, Matsumura, H, Hokao, R. | | Deposit date: | 2019-06-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural analysis of hexameric structure of glycerol kinase from Thermococcus kodakaraeinsis KOD1
To Be Published
|
|
6KVQ
 
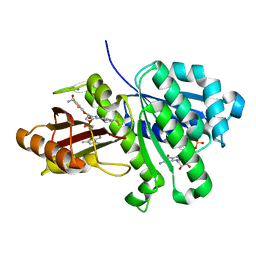 | | S. aureus FtsZ in complex with BOFP (compound 3) | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
6K79
 
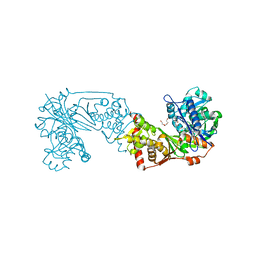 | | Glycerol kinase form Thermococcus kodakarensis, complex structure with substrate. | | Descriptor: | GLYCEROL, Glycerol kinase, TRIETHYLENE GLYCOL | | Authors: | Koga, Y, Angkawidjaja, C, Matsumura, H, Hokao, R. | | Deposit date: | 2019-06-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural analysis of hexameric structure of glycerol kinase from Thermococcus kodakaraeinsis KOD1
To Be Published
|
|
6KVP
 
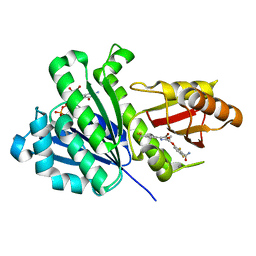 | | S. aureus FtsZ in complex with 3-(1-(5-bromo-4-(4-(trifluoromethyl)phenyl)oxazol-2-yl)ethoxy)-2,6-difluorobenzamide (compound 2) | | Descriptor: | 3-[(1R)-1-[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]ethoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
3WR7
 
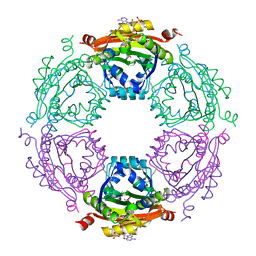 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
3VKW
 
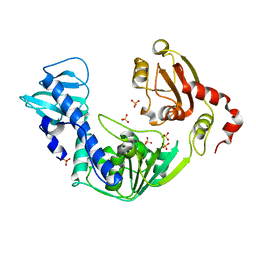 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
3WYQ
 
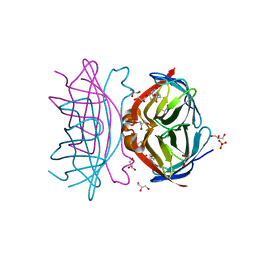 | | Crystal structure of the low-immunogenic core streptavidin mutant LISA-314 (Y22S/Y83S/R84K/E101D/R103K/E116N) at 1.0 A resolution | | Descriptor: | BIOTIN, GLYCEROL, SULFATE ION, ... | | Authors: | Kawato, T, Mizohata, E, Meshizuka, T, Doi, H, Kawamura, T, Matsumura, H, Yumura, K, Tsumoto, K, Kodama, T, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of streptavidin mutant with low immunogenicity.
J.Biosci.Bioeng., 119, 2015
|
|
4YSP
 
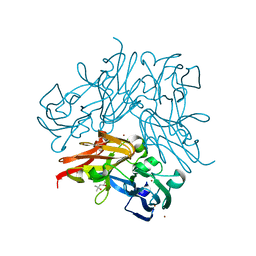 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
