5CTQ
 
 | |
5CTR
 
 | |
1WS0
 
 | |
1WS1
 
 | |
3OQC
 
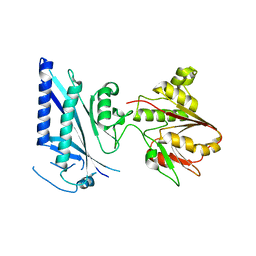 | |
3E21
 
 | |
1ICE
 
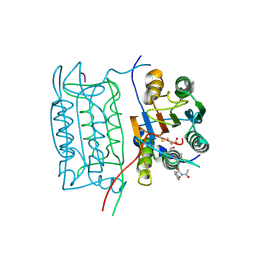 | | STRUCTURE AND MECHANISM OF INTERLEUKIN-1BETA CONVERTING ENZYME | | Descriptor: | INTERLEUKIN-1 BETA CONVERTING ENZYME, TETRAPEPTIDE ALDEHYDE | | Authors: | Wilson, K.P, Griffith, J.P, Kim, E.E, Navia, M.A. | | Deposit date: | 1994-09-29 | | Release date: | 1995-07-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of interleukin-1 beta converting enzyme.
Nature, 370, 1994
|
|
5Y7J
 
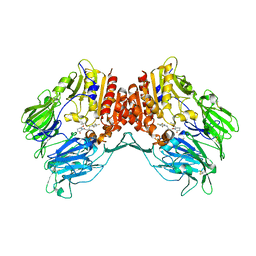 | | Crystal structure of human DPP4 in complex with inhibitor2 | | Descriptor: | (S)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7H
 
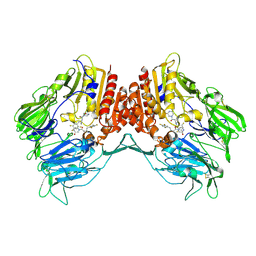 | | Crystal structure of human DPP4 in complex with inhibitor3 | | Descriptor: | (R)-4-(3-amino-4-(2,4,5-trifluorophenyl)butanoyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7K
 
 | | Crystal structure of human DPP4 in complex with inhibitor1 | | Descriptor: | (R)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazine-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5X3S
 
 | |
5XDA
 
 | |
3CMD
 
 | | Crystal structure of peptide deformylase from VRE-E.faecium | | Descriptor: | FE (III) ION, MALONATE ION, Peptide deformylase, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2008-03-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into the antibacterial drug design and architectural mechanism of peptide recognition from the E. faecium peptide deformylase structure.
Proteins, 74, 2009
|
|
2FI7
 
 | |
2RLN
 
 | |
3W8L
 
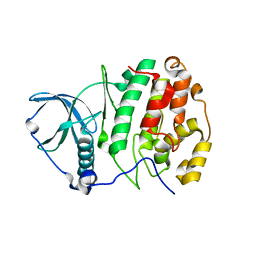 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2GX2
 
 | | Crystal structural and functional analysis of GFP-like fluorescent protein Dronpa | | Descriptor: | MAGNESIUM ION, fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.-H, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the photoswitchable fluorescent protein Dronpa-C62S
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2GX0
 
 | |
8ENJ
 
 | |
8GUW
 
 | |
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
3G6N
 
 | | Crystal structure of an EfPDF complex with Met-Ala-Ser | | Descriptor: | FE (III) ION, Peptide deformylase, SODIUM ION, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3IHC
 
 | |
3IIG
 
 | |
3ILB
 
 | |
