4LT5
 
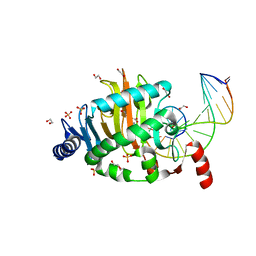 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | Authors: | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
8W9E
 
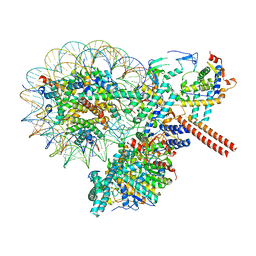 | |
8W9C
 
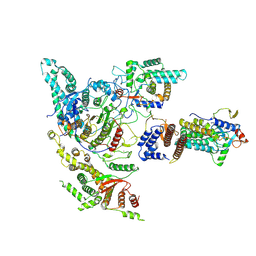 | |
8W9D
 
 | |
6D2Q
 
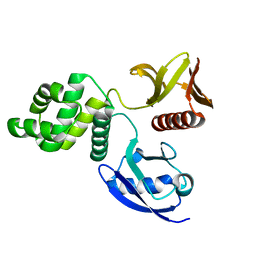 | | Crystal structure of the FERM domain of zebrafish FARP1 | | Descriptor: | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | | Authors: | Kuo, Y.C, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
4R2E
 
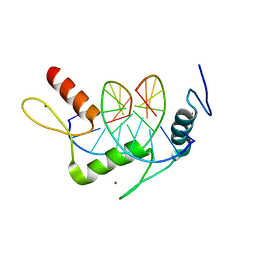 | | Wilms Tumor Protein (WT1) zinc fingers in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2A
 
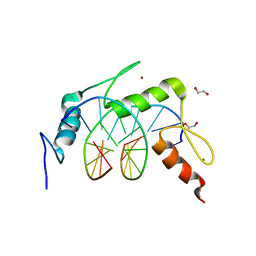 | | Egr1/Zif268 zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
6D21
 
 | |
4R2D
 
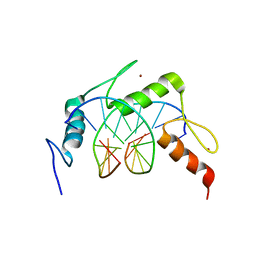 | | Egr1/Zif268 zinc fingers in complex with formylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2C
 
 | | Egr1/Zif268 zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
3EHQ
 
 | | Crystal Structure of Human Osteoclast Stimulating Factor | | Descriptor: | 1,2-ETHANEDIOL, Osteoclast-stimulating factor 1 | | Authors: | Tong, S, Zhou, H, Gao, Y, Zhu, Z, Zhang, X, Teng, M, Niu, L. | | Deposit date: | 2008-09-14 | | Release date: | 2009-08-04 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of human osteoclast stimulating factor
Proteins, 75, 2009
|
|
4R2S
 
 | | Wilms Tumor Protein (WT1) Q369P zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4M9E
 
 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-14 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
3LOI
 
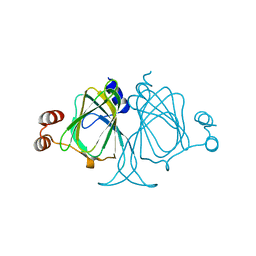 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in the apo and GDP-bound forms | | Descriptor: | Putative uncharacterized protein | | Authors: | Zou, C.Z, Du, Y, He, Y.-X, Saren, G, Zhang, X, Chen, Y, Zhang, S.-C. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
8EW2
 
 | |
8F49
 
 | |
8EW0
 
 | |
8F4L
 
 | |
8F7Y
 
 | |
3IG3
 
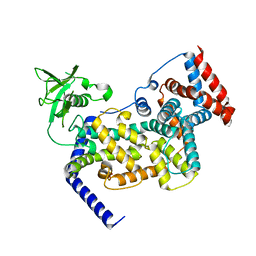 | | Crystal structure of mouse Plexin A3 intracellular domain | | Descriptor: | GLYCEROL, Plxna3 protein | | Authors: | He, H, Zhang, X. | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2IPA
 
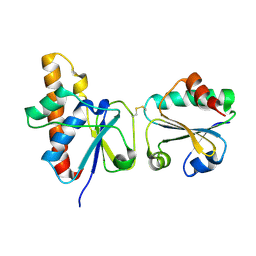 | | solution structure of Trx-ArsC complex | | Descriptor: | Protein arsC, Thioredoxin | | Authors: | Jin, C, Hu, Y, Li, Y, Zhang, X. | | Deposit date: | 2006-10-12 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
8GWA
 
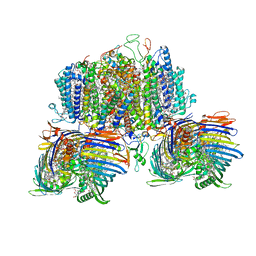 | | Structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium.
J Integr Plant Biol, 65, 2023
|
|
2J48
 
 | |
3JB5
 
 | |
7M0R
 
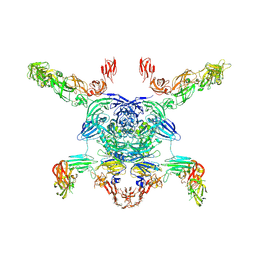 | | Cryo-EM structure of the Sema3A/PlexinA4/Neuropilin 1 complex | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A4, ... | | Authors: | Lu, D, Shang, G, He, X, Bai, X, Zhang, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex.
Nat Commun, 12, 2021
|
|
