6AO5
 
 | | Crystal structure of human MST2 in complex with SAV1 SARAH domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein salvador homolog 1, ... | | Authors: | Tomchick, D.R, Luo, X, Ni, L. | | Deposit date: | 2017-08-15 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK.
Elife, 6, 2017
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7UUH
 
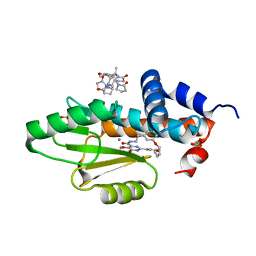 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001034732-2 (cis-form) | | Descriptor: | (1P,18Z)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2(28),5,11(27),12,14,18,22,24-nonaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7UMR
 
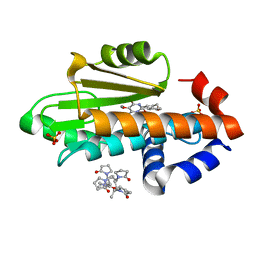 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001034732-1 (trans-form) | | Descriptor: | (1P,18Z)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2(28),5,11(27),12,14,18,22,24-nonaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-04-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7VVC
 
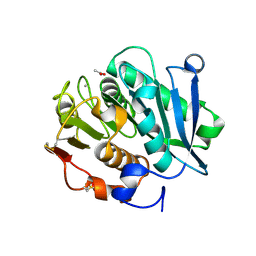 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7VQ6
 
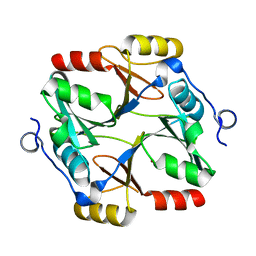 | | Structure of a specialized glyoxalase from Gossypium hirsutum | | Descriptor: | Lactoylglutathione lyase, NICKEL (II) ION | | Authors: | Li, H, Hu, Y.M, Dai, L.H, Chen, C.C, Huang, J.W, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure and biochemical analysis of the specialized deoxynivalenol-detoxifying glyoxalase SPG from Gossypium hirsutum.
Int.J.Biol.Macromol., 200, 2022
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
3SMQ
 
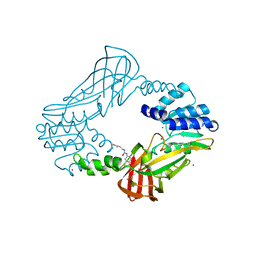 | | Crystal structure of protein arginine methyltransferase 3 | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | Authors: | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
7LP8
 
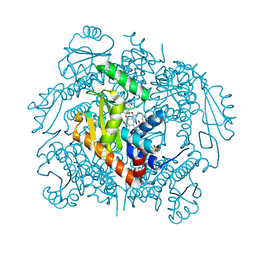 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7LP7
 
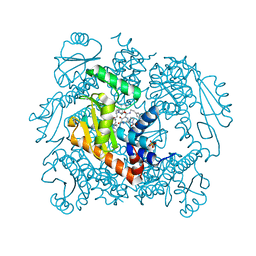 | | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7CMA
 
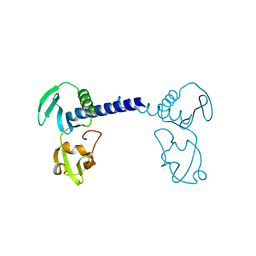 | | Structure of A151R from African swine fever virus Georgia | | Descriptor: | A151R, ZINC ION | | Authors: | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | Deposit date: | 2020-07-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7ML8
 
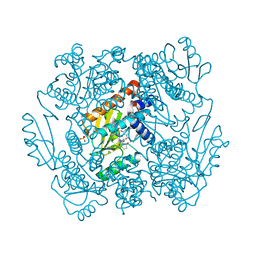 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ001023038 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxamide, GLUTAMIC ACID, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-04-27 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with St Jude compound AC067-19
To Be Published
|
|
7MX0
 
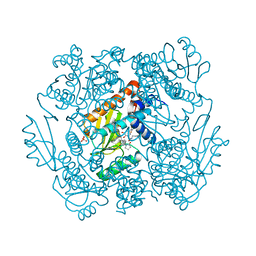 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988558 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-17 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988558
To Be Published
|
|
7MY5
 
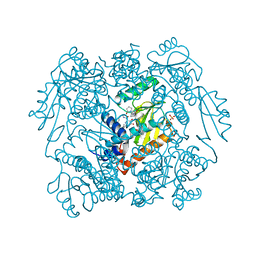 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-20 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503
To Be Published
|
|
7EPZ
 
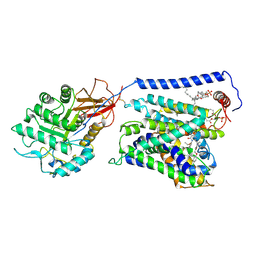 | | Overall structure of Erastin-bound xCT-4F2hc complex | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-[(1S)-1-[4-[2-(4-chloranylphenoxy)ethanoyl]piperazin-1-yl]ethyl]-3-(2-ethoxyphenyl)quinazolin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis.
Cell Res., 32, 2022
|
|
7CWQ
 
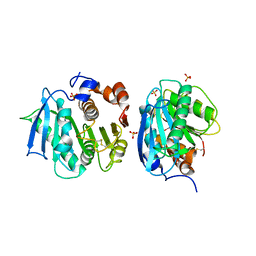 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
7K77
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025
To Be Published
|
|
7KNR
 
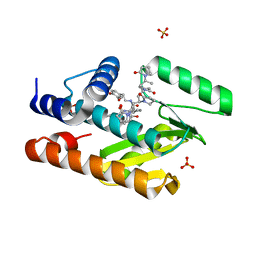 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558
To Be Published
|
|
7KNY
 
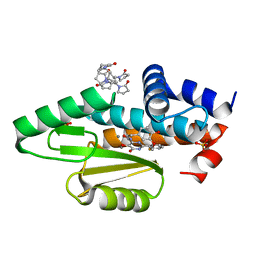 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-06 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528
To Be Published
|
|
6BPH
 
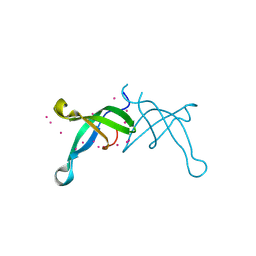 | | Crystal structure of the chromodomain of RBBP1 | | Descriptor: | AT-rich interactive domain-containing protein 4A, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of chromo barrel domain of RBBP1.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
7CY0
 
 | | Crystal structure of S185H mutant PET hydrolase from Ideonella sakaiensis | | Descriptor: | ACETIC ACID, Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
3PKI
 
 | | Human SIRT6 crystal structure in complex with ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
