2KZT
 
 | | Structure of the Tandem MA-3 Region of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Waters, L.C, Strong, S.L, Oka, O, Muskett, F.W, Veverka, V, Banerjee, S, Schmedt, T, Henry, A.J, Klempnauer, K.H, Carr, M.D. | | Deposit date: | 2010-06-24 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor
J.Biol.Chem., 286, 2011
|
|
2L73
 
 | |
2C1A
 
 | | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2C1B
 
 | | Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-(4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline | | Descriptor: | (4R,2S)-5'-(4-(4-CHLOROBENZYLOXY)PYRROLIDIN-2-YLMETHANESULFONYL)ISOQUINOLINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
2M5S
 
 | | High-resolution NMR structure and cryo-EM imaging support multiple functional roles for the accessory I-domain of phage P22 coat protein | | Descriptor: | Coat protein | | Authors: | Rizzo, A.A, Suhanovsky, M.M, Baker, M.L, Fraser, L.C.R, Jones, L.M, Rempel, D.L, Gross, M.L, Chiu, W, Alexandrescu, A.T, Teschke, C.M. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling.
Structure, 22, 2014
|
|
2H5Y
 
 | | Crystallographic structure of the Molybdate-Binding Protein of Xanthomonas citri at 1.7 Ang resolution bound to molybdate | | Descriptor: | MOLYBDATE ION, Molybdate-binding periplasmic protein, SULFATE ION | | Authors: | Balan, A, Santacruz, C.P, Ferreira, L.C.S, Barbosa, J.A.R.G. | | Deposit date: | 2006-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallization, data collection and phasing of the molybdate-binding protein of the phytopathogen Xanthomonas axonopodis pv. citri
Acta Crystallogr.,Sect.F, 62, 2006
|
|
6QSR
 
 | | The Dehydratase Heterocomplex ApeI:P from Xenorhabdus doucetiae | | Descriptor: | AMP-dependent synthetase and ligase, Beta-hydroxyacyl-(Acyl-carrier-protein) dehydratase FabA/FabZ, SODIUM ION | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
7KJU
 
 | | Cgi121-tRNA complex | | Descriptor: | MAGNESIUM ION, RNA (75-MER) | | Authors: | Ceccarelli, D.F, Beenstock, J, Wan, L.C.K, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
6UY4
 
 | | Crystal structure of dihydroorotate dehydrogenase from Schistosoma mansoni | | Descriptor: | 2-[(4-fluorophenyl)amino]-3-hydroxynaphthalene-1,4-dione, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mori, R.M, Zapata, L.C.C, Nonato, M.C. | | Deposit date: | 2019-11-11 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural basis for the function and inhibition of dihydroorotate dehydrogenase from Schistosoma mansoni.
Febs J., 288, 2021
|
|
6NDQ
 
 | |
6QSP
 
 | | Ketosynthase (ApeO) in Complex with its Chain Length Factor (ApeC) from Xenorhabdus doucetiae | | Descriptor: | Beta-ketoacyl synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grammbitter, G.L.C, Schmalhofer, M, Groll, M, Bode, H. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments.
J.Am.Chem.Soc., 141, 2019
|
|
6SM4
 
 | | AntF (apo): type II PKS acyl-carrier protein | | Descriptor: | Acyl carrier protein | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SMD
 
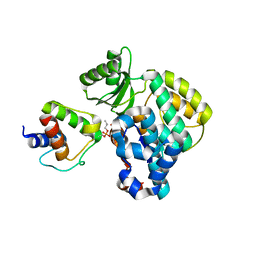 | | PlMCAT:AntF (holo): type II PKS acyl-carrier protein in complex with its malonyl-transacylase | | Descriptor: | Acyl carrier protein, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SM6
 
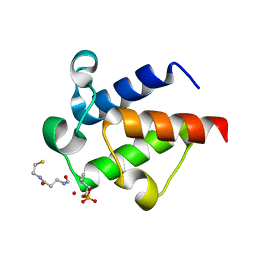 | | AntF (holo): type II PKS acyl-carrier protein | | Descriptor: | Acyl carrier protein | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SMO
 
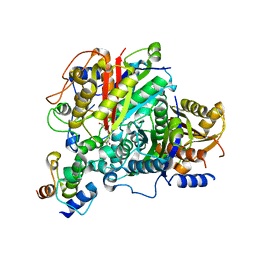 | | AntDE:AntF (apo): type II PKS acyl-carrier protein in complex with its ketosynthase bound to the hexaketide | | Descriptor: | 3,5,7,9,11-pentakis(oxidanylidene)dodecanal, Acyl carrier protein, Ketoacyl_synth_N domain-containing protein, ... | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
6SMP
 
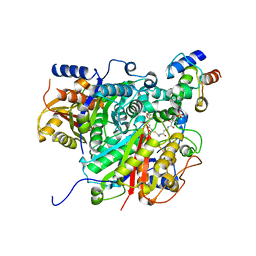 | | AntDE:AntF (holo): type II PKS acyl-carrier protein in complex with its ketosynthase bound to the hexaketide | | Descriptor: | (2~{R})-3,3-dimethyl-2-oxidanyl-~{N}-[3-oxidanylidene-3-[2-[(1~{R},4~{Z},6~{Z},8~{Z})-1,5,7,9-tetrakis(oxidanyl)-3,11-bis(oxidanylidene)dodeca-4,6,8-trienyl]sulfanylethylamino]propyl]-4-[tris(oxidanyl)-$l^{5}-phosphanyl]oxy-butanamide, Acyl carrier protein, Ketoacyl_synth_N domain-containing protein, ... | | Authors: | Braeuer, A, Zhou, Q, Grammbitter, G.L.C, Schmalhofer, M, Ruehl, M, Kaila, V.R.I, Bode, H, Groll, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Nat.Chem., 12, 2020
|
|
1SGW
 
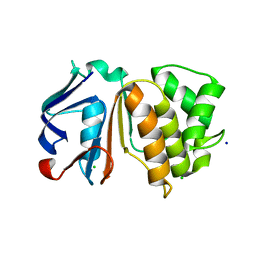 | | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001 | | Descriptor: | CHLORIDE ION, SODIUM ION, putative ABC transporter | | Authors: | Liu, Z.J, Tempel, W, Shah, A, Chen, L, Lee, D, Kelley, L.-L.C, Dillard, B.D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001
To be Published
|
|
1XI3
 
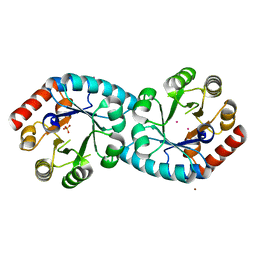 | | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Thiamine phosphate pyrophosphorylase, ... | | Authors: | Zhou, W, Chen, L, Tempel, W, Liu, Z.-J, Habel, J, Lee, D, Lin, D, Chang, S.-H, Dillard, B.D, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Kelley, L.-L.C, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001
To be published
|
|
1XX7
 
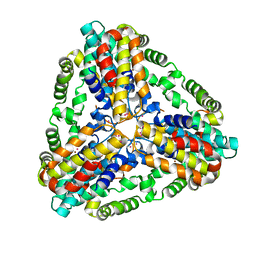 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001 | | Descriptor: | NICKEL (II) ION, UNKNOWN ATOM OR ION, oxetanocin-like protein | | Authors: | Chen, L, Tempel, W, Habel, J, Zhou, W, Nguyen, D, Chang, S.-H, Lee, D, Kelley, L.-L.C, Dillard, B.D, Liu, Z.-J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-11-04 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-403030-001
To be published
|
|
1YBZ
 
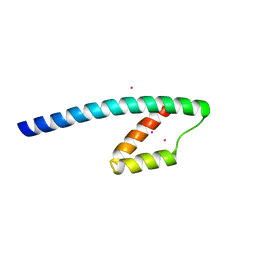 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-1581948-001 | | Descriptor: | UNKNOWN ATOM OR ION, chorismate mutase | | Authors: | Lee, D, Chen, L, Nguyen, D, Dillard, B.D, Tempel, W, Habel, J, Zhou, W, Chang, S.-H, Kelley, L.-L.C, Liu, Z.-J, Lin, D, Zhang, H, Praissman, J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-1581948-001
To be published
|
|
2VTM
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
5IGN
 
 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
2VU3
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTL
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-phenyl-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTT
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-difluorophenyl)carbonyl]amino}-N-[(3S)-piperidin-3-yl]-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
