8YDQ
 
 | |
8YDP
 
 | |
8YDT
 
 | |
8YDW
 
 | |
8YDX
 
 | |
8YDS
 
 | |
3CTH
 
 | |
3CTJ
 
 | |
5D1J
 
 | |
4P58
 
 | | Crystal structure of mouse comt bound to an inhibitor | | Descriptor: | 1',3'-dimethyl-1H,1'H-3,4'-bipyrazole, Catechol O-methyltransferase | | Authors: | Lanier, M. | | Deposit date: | 2014-03-15 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A fragment-based approach to identifying S-adenosyl-l-methionine -competitive inhibitors of catechol O-methyl transferase (COMT).
J.Med.Chem., 57, 2014
|
|
5B71
 
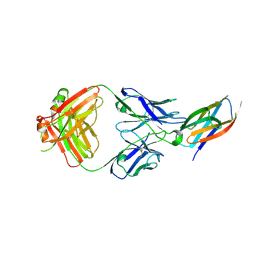 | | Crystal structure of complement C5 in complex with SKY59 | | Descriptor: | Complement C5 beta chain, SKY59 Fab heavy chain, SKY59 Fab light chain | | Authors: | Irie, M, Shimizu, Y, Sampei, Z, Fukuzawa, T. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases.
Sci Rep, 7, 2017
|
|
8JH0
 
 | | Crystal structure of the light-driven sodium pump IaNaR | | Descriptor: | RETINAL, Xanthorhodopsin | | Authors: | Hashimoto, T, Kato, K, Tanaka, Y, Yao, M, Kikukawa, T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Multistep conformational changes leading to the gate opening of light-driven sodium pump rhodopsin.
J.Biol.Chem., 299, 2023
|
|
8DD5
 
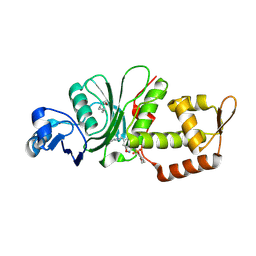 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | Descriptor: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Greasley, S.E, Johnson, E, Brodsky, O. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
8SKQ
 
 | |
8YUQ
 
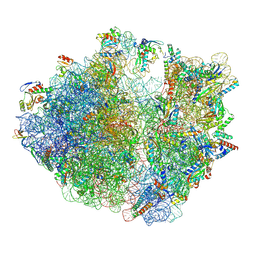
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and dA4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUP
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and A4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-28 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUR
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and Am4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUS
 
 | | E. coli 70S ribosome complexed with P.putida tRNAIle2 and A(F)4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUO
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 at the A-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8JYM
 
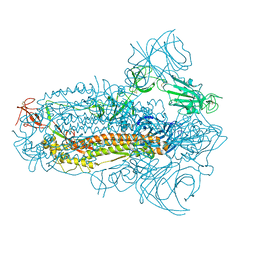 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8JYK
 
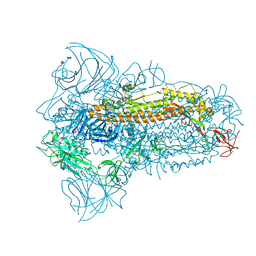 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8JYP
 
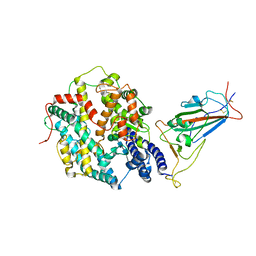 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
3F82
 
 | |
8JYN
 
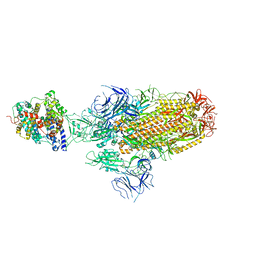 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8JYO
 
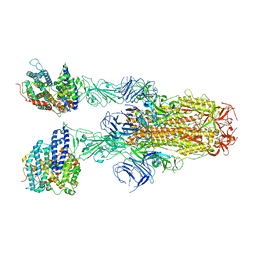 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
