2BX9
 
 | | Crystal structure of B.subtilis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | TRYPTOPHAN RNA-BINDING ATTENUATOR PROTEIN-INHIBITORY PROTEIN, ZINC ION | | Authors: | Shevtsov, M.B, Chen, Y, Gollnick, P, Antson, A.A. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Anti-Trap Protein, an Antagonist of Trap/RNA Interaction.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4GM8
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-102 | | Descriptor: | MM-102, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
4G43
 
 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
4GM9
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-401 | | Descriptor: | MM-401, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Liu, L, Dou, Y, Lei, M, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human WD repeat domain 5 with compound MM-401
To be Published
|
|
6UJB
 
 | | Integrin alpha-v beta-8 in complex with the Fabs C6D4 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6D4 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
4F11
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor GBR2 | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Geng, Y, Xiong, D, Mosyak, L, Malito, D.L, Kniazeff, J, Chen, Y, Burmakina, S, Quick, M, Bush, M, Javitch, J.A, Pin, J.-P, Fan, Q.R. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2.
Nat.Neurosci., 15, 2012
|
|
6W4S
 
 | | Structure of apo human ferroportin in lipid nanodisc | | Descriptor: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6WBV
 
 | | Structure of human ferroportin bound to hepcidin and cobalt in lipid nanodisc | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, COBALT (II) ION, Fab45D8 Heavy Chain, ... | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6W4V
 
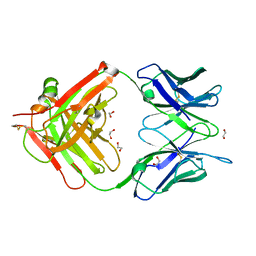 | | Structure of anti-ferroportin Fab45D8 | | Descriptor: | 1,2-ETHANEDIOL, Fab45D8 Heavy Chain, Fab45D8 Light Chain | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6E1N
 
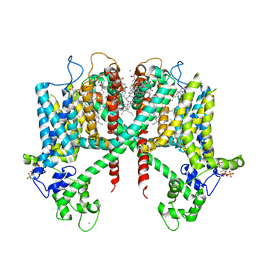 | | Structure of AtTPC1(DDE) in state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1K
 
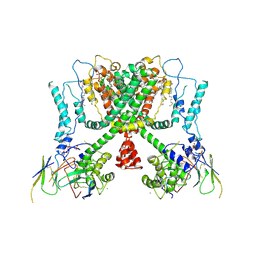 | | Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3QJT
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
2ASQ
 
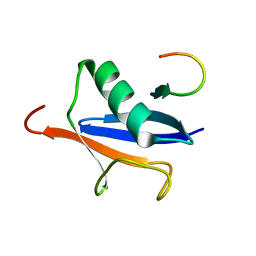 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
2LJV
 
 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
2B6E
 
 | | X-Ray Crystal Structure of Protein HI1161 from Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR63. | | Descriptor: | ACETIC ACID, Hypothetical UPF0152 protein HI1161 | | Authors: | Kuzin, A.P, Benach, J, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Cunningham, K.E, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-01 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the different crystal form of the hypothetical UPF0152 protein HI1161: NESG target IR63
TO BE PUBLISHED
|
|
2NPP
 
 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
1MVO
 
 | | Crystal structure of the PhoP receiver domain from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, PhoP response regulator, SODIUM ION | | Authors: | Birck, C, Chen, Y, Hulett, F.M, Samama, J.P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-09-26 | | Release date: | 2002-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Phosphorylation Domain in PhoP Reveals a Functional Tandem Association Mediated by an Asymmetric Interface
J.BACTERIOL., 185, 2003
|
|
1ZBR
 
 | | Crystal Structure of the Putative Arginine Deiminase from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR3 | | Descriptor: | conserved hypothetical protein | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Putative Arginine Deiminase from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR3
To be Published
|
|
3QJR
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
4GM3
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-101 | | Descriptor: | MM-101, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.393 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
4GP4
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
3QJS
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJQ
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJU
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
4GP8
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W+T231F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
