6EXT
 
 | |
6EXU
 
 | |
6FCV
 
 | | Structure of the human DDB1-CSA complex | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-8 | | Authors: | Meulenbroek, E.M, Pannu, N.S. | | Deposit date: | 2017-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A.
Nat Commun, 9, 2018
|
|
6FGY
 
 | | Crystal Structure of Human BACE-1 in Complex with amino-1,4-oxazine compound 4 | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R})-5-azanyl-3-methyl-2,6-dihydro-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.-M, Bourgier, E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of amino-1,4-oxazines as potent BACE-1 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
8P08
 
 | | Crystal structure of human CLK1 in complex with Leucettinib-21 | | Descriptor: | (4~{Z})-4-(1,3-benzothiazol-6-ylmethylidene)-2-[[(2~{R})-1-methoxy-4-methyl-pentan-2-yl]amino]-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical, Biochemical, Cellular, and Physiological Characterization of Leucettinib-21, a Down Syndrome and Alzheimer's Disease Drug Candidate.
J.Med.Chem., 66, 2023
|
|
8B0Q
 
 | | Deinococcus radiodurans UvrC C-terminal half | | Descriptor: | UvrABC system protein C | | Authors: | Timmins, J, Stelter, M. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.
Nucleic Acids Res., 51, 2023
|
|
7BLV
 
 | |
7BM0
 
 | | Crystal structure of the tick-borne encephalitis virus NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase domain, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anindita, P.D, Grinkevich, P, Franta, Z. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
6GK8
 
 | | Crystal structure of anti-tau antibody dmCBTAU-28.1, double mutant (S32R, E35K) of CBTAU-28.1, in complex with Tau peptide A7731 (residues 52-71) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-28.1(S32R;E35K), TAU PEPTIDE A7731 (RESIDUES 52-71) | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
8DQV
 
 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7KQL
 
 | | Anti-Tim3 antibody Fab complex | | Descriptor: | GLYCEROL, Hepatitis A virus cellular receptor 2, Tim3.18 Fab heavy chain, ... | | Authors: | Deng, X.A, West, S.M, Strop, P. | | Deposit date: | 2020-11-16 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Tim-3 mediates T cell trogocytosis to limit antitumor immunity.
J.Clin.Invest., 132, 2022
|
|
6GK7
 
 | | Crystal structure of anti-tau antibody dmCBTAU-27.1, double mutant (S31Y, T100I) of CBTAU-27.1, in complex with Tau peptide A8119B (residues 299-318) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-27.1(S31Y,T100I), HUMAN TAU PEPTIDE A8119 RESIDUES 299-318 | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
8B7Z
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Palm, G.J, Hinrichs, W. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7R
 
 | | Bacterial chalcone isomerase with taxifolin chalcone | | Descriptor: | (Z)-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-1-[2,4,6-tris(oxidanyl)phenyl]prop-2-en-1-one, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7U
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
5ENW
 
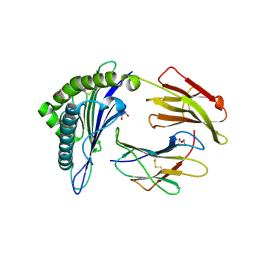 | | Structure of HLA-A2:01 with peptide G9L | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Remesh, S.G, Zajonc, D.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unconventional Peptide Presentation by Major Histocompatibility Complex (MHC) Class I Allele HLA-A*02:01: BREAKING CONFINEMENT.
J. Biol. Chem., 292, 2017
|
|
7PV1
 
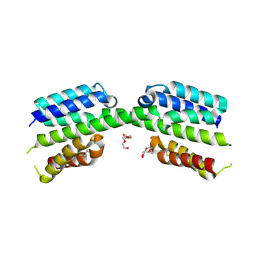 | |
7PUZ
 
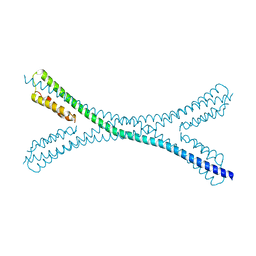 | |
7PV0
 
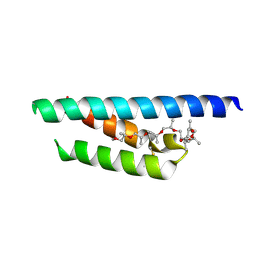 | | Crystal structure of a Mic60-Mic19 fusion protein | | Descriptor: | MICOS complex subunit MIC60,MICOS complex subunit MIC60-MIC19,Mic60-Mic19, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Funck, K, Bock-Bierbaum, T, Daumke, O. | | Deposit date: | 2021-10-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into crista junction formation by the Mic60-Mic19 complex.
Sci Adv, 8, 2022
|
|
5F7D
 
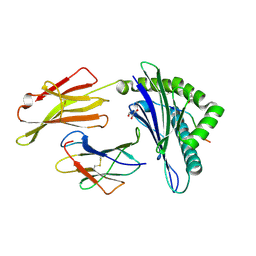 | | Structure of HLA-A2:01 with peptide G11N | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Zajonc, D.M, Remesh, S.G. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unconventional Peptide Presentation by Major Histocompatibility Complex (MHC) Class I Allele HLA-A*02:01: BREAKING CONFINEMENT.
J. Biol. Chem., 292, 2017
|
|
6EZV
 
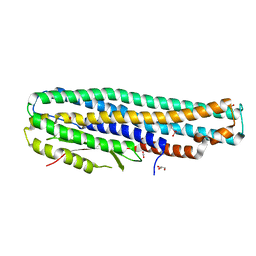 | | The cytotoxin MakA from Vibrio cholerae | | Descriptor: | ACETATE ION, CACODYLATE ION, GLYCEROL, ... | | Authors: | Persson, K, Dongre, M, Wai, S.N. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flagella-mediated secretion of a novelVibrio choleraecytotoxin affecting both vertebrate and invertebrate hosts.
Commun Biol, 1, 2018
|
|
6YWD
 
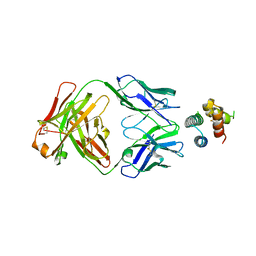 | | De novo designed protein 4H_01 in complex with Mota antibody | | Descriptor: | Antibody Mota, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
6YWC
 
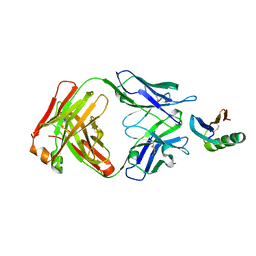 | | De novo designed protein 4E1H_95 in complex with 101F antibody | | Descriptor: | Antibody 101F, Heavy Chain, light chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
5F9J
 
 | | Structure of HLA-A2:01 with peptide Y9L | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Remesh, S.G, Zajonc, D.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Unconventional Peptide Presentation by Major Histocompatibility Complex (MHC) Class I Allele HLA-A*02:01: BREAKING CONFINEMENT.
J. Biol. Chem., 292, 2017
|
|
5FA3
 
 | | Structure of HLA-A2:01 with peptide G9V | | Descriptor: | Beta-2-microglobulin, G9V, GLYCEROL, ... | | Authors: | Zajonc, D.M, Remesh, S.G. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Unconventional Peptide Presentation by Major Histocompatibility Complex (MHC) Class I Allele HLA-A*02:01: BREAKING CONFINEMENT.
J. Biol. Chem., 292, 2017
|
|
