8HJH
 
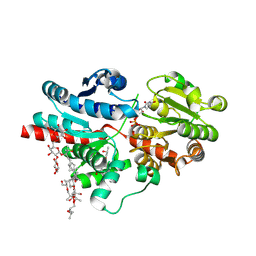 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the catalytic selectivity of SgUGT94-289-3 towards mogrosides
To Be Published
|
|
8HJK
 
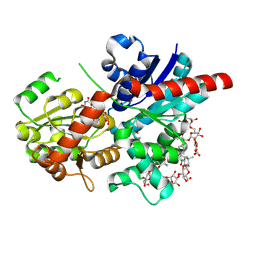 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 1 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural insights into the catalytic selectivity of SgUGT94-289-3 towards mogrosides
To Be Published
|
|
8HJL
 
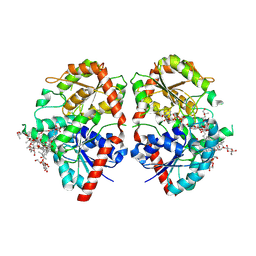 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3E | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(3S,8S,9R,10S,11S,13R,14S,17S)-17-[(2S,5R)-5-[(2S,3R,4S,5R,6S)-6-(hydroxymethyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insights into the catalytic selectivity of SgUGT94-289-3 towards mogrosides
To Be Published
|
|
6P3Q
 
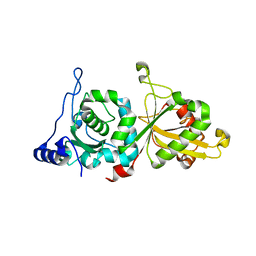 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
8I2H
 
 | | Follicle stimulating hormone receptor | | Descriptor: | Follicle-stimulating hormone receptor | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
8I2G
 
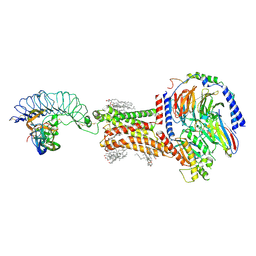 | | FSHR-Follicle stimulating hormone-compound 716340-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
1ET1
 
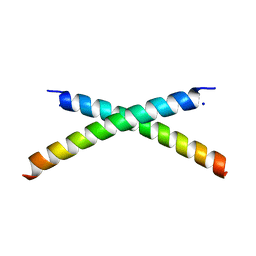 | | CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESOLUTION | | Descriptor: | PARATHYROID HORMONE, SODIUM ION | | Authors: | Jin, L, Briggs, S.L, Chandrasekhar, S, Chirgadze, N.Y, Clawson, D.K, Schevitz, R.W, Smiley, D.L, Tashjian, A.H, Zhang, F. | | Deposit date: | 2000-04-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of human parathyroid hormone 1-34 at 0.9-A resolution.
J.Biol.Chem., 275, 2000
|
|
3NIV
 
 | |
3NND
 
 | |
3NPK
 
 | |
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
6QNS
 
 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B mutant I1248W/V1249W in complex with human synaptotagmin 1 and GD1a receptors | | Descriptor: | Botulinum neurotoxin type B, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Synaptotagmin-1 | | Authors: | Masuyer, G, Yin, L, Zhang, S, Miyashita, S.I, Dong, M, Stenmark, P. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy.
Plos Biol., 18, 2020
|
|
3PAN
 
 | |
3PAO
 
 | |
3PU6
 
 | |
3Q34
 
 | |
3NAS
 
 | |
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
7RHB
 
 | |
7RHD
 
 | | darkmRuby M94T/F96Y mutant at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby M94T/F96Y mutant | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
8BF9
 
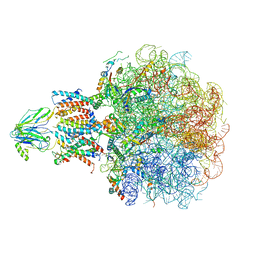 | | Molecular view of ER membrane remodeling by the Sec61/TRAP translocon. | | Descriptor: | 60S ribosomal protein L39, Large ribosomal subunit protein eL31, Large ribosomal subunit protein eL38, ... | | Authors: | Karki, S, Javanainen, M, Tranter, D, Rehan, S, Huiskonen, J, Happonen, L, Paavilainen, V. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular view of ER membrane remodeling by the Sec61/TRAP translocon.
Embo Rep., 24, 2023
|
|
7RHC
 
 | | A new fluorescent protein darkmRuby at pH 9.0 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
3PBK
 
 | |
3PBM
 
 | |
2G59
 
 | |
