1Q0X
 
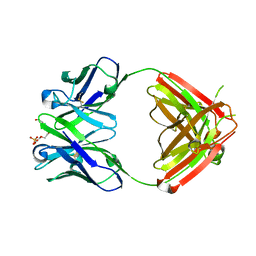 | | Anti-morphine Antibody 9B1 Unliganded Form | | Descriptor: | Fab 9B1, heavy chain, light chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
1RYO
 
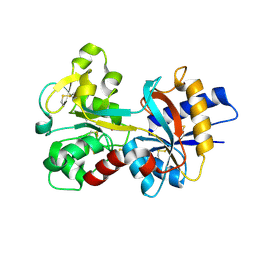 | | Human serum transferrin, N-lobe bound with oxalate | | Descriptor: | FE (III) ION, OXALATE ION, Serotransferrin | | Authors: | Halbrooks, P.J, Mason, A.B, Adams, T.E, Briggs, S.K, Everse, S.J. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The oxalate effect on release of iron from human serum transferrin explained.
J.Mol.Biol., 339, 2004
|
|
1RRJ
 
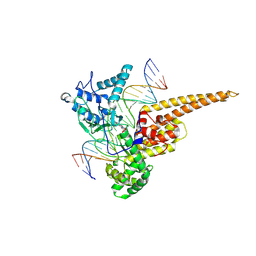 | | Structural Mechanisms of Camptothecin Resistance by Mutations in Human Topoisomerase I | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T*GP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Chrencik, J.E, Staker, B.L, Burgin, A.B, Stewart, L, Redinbo, M.R. | | Deposit date: | 2003-12-08 | | Release date: | 2004-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of camptothecin resistance by human topoisomerase I mutations
J.Mol.Biol., 339, 2004
|
|
4B29
 
 | | Crystal structures of DMSP lyases RdDddP and RnDddQII | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DIMETHYLSULFONIOPROPIONATE LYASE, ... | | Authors: | Hehemann, J.H, Law, A, Redecke, L, Boraston, A.B. | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structures of Dmsp Lyases Rddddp and Rndddqii
To be Published
|
|
4C22
 
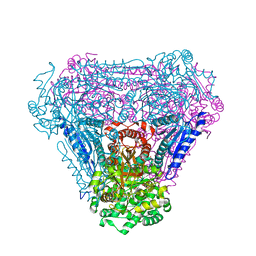 | | L-Fucose Isomerase In Complex With Fuculose | | Descriptor: | 1,2-ETHANEDIOL, L-FUCOSE ISOMERASE, L-Fuculose open form, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4BQ4
 
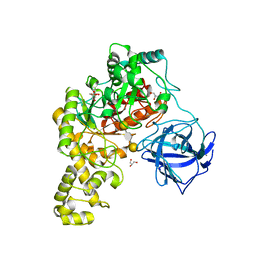 | | Structural analysis of an exo-beta-agarase | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, B-AGARASE, CALCIUM ION, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
4C1S
 
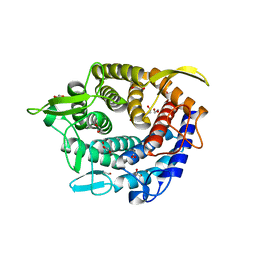 | | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 76 MANNOSIDASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4BQ5
 
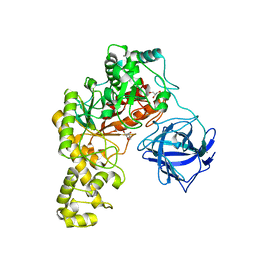 | | Structural analysis of an exo-beta-agarase | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
1SI1
 
 | | Crystal Structure of Mannheimia haemolytica Ferric iron-Binding Protein A in an open conformation | | Descriptor: | FE (III) ION, iron binding protein FbpA | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, Snell, G, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens.
J.Bacteriol., 186, 2004
|
|
1SEU
 
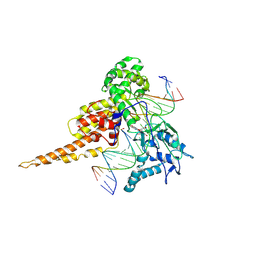 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Indolocarbazole SA315F and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 2,10-DIHYDROXY-12-(BETA-D-GLUCOPYRANOSYL)-6,7,12,13-TETRAHYDROINDOLO[2,3-A]PYRROLO[3,4-C]CARBAZOLE-5,7-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-02-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1SC7
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Indenoisoquinoline MJ-II-38 and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 4-(5,11-DIOXO-5H-INDENO[1,2-C]ISOQUINOLIN-6(11H)-YL)BUTANOATE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-02-11 | | Release date: | 2005-04-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
7JNB
 
 | |
7JS4
 
 | |
7JND
 
 | |
7JNF
 
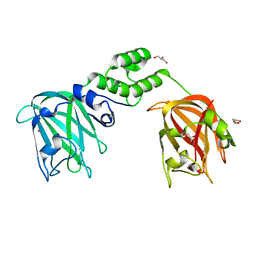 | |
7JWF
 
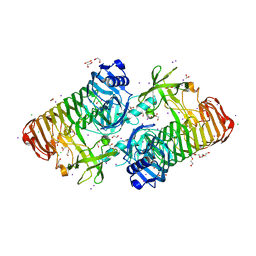 | | Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2020-08-25 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structure of a family 110 glycoside hydrolase provides insight into the hydrolysis of alpha-1,3-galactosidic linkages in lambda-carrageenan and blood group antigens.
J.Biol.Chem., 295, 2020
|
|
7JRM
 
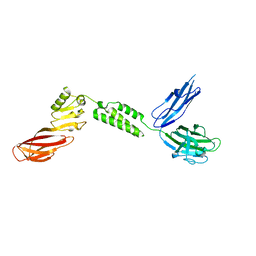 | |
7JRL
 
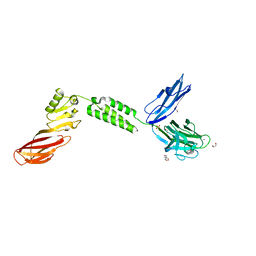 | | The structure of CBM51-2 in complex with GlcNAc and INT domains from Clostridium perfringens ZmpB | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-08-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecturally complex O -glycopeptidases are customized for mucin recognition and hydrolysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JW4
 
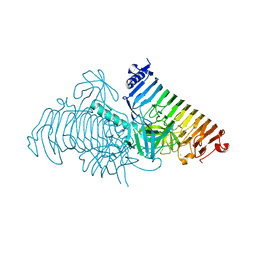 | | Crystal structure of PdGH110B in complex with D-galactose | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 110, NICKEL (II) ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | The structure of a family 110 glycoside hydrolase provides insight into the hydrolysis of alpha-1,3-galactosidic linkages in lambda-carrageenan and blood group antigens.
J.Biol.Chem., 295, 2020
|
|
7JTV
 
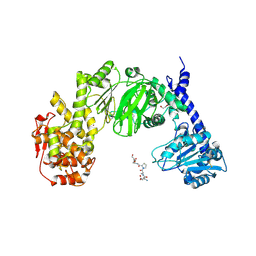 | |
7JFS
 
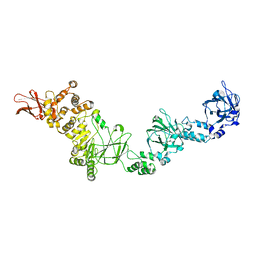 | |
7K53
 
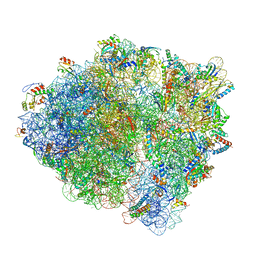 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K54
 
 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
