4XSB
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
7SP2
 
 | | Structure of PLS A-domain (residues 391-656; 513-518 deletion mutant) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Plasmin Sensitive Protein Pls | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of PLS A-domain (residues 391-65) from Staphylococcus aureus
Not Published
|
|
5J26
 
 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
4QGT
 
 | | The Crystal Structure of Human IgG Fc Domain with Enhanced Aromatic Sequon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kong, L, Connelly, S.C, Wilson, I.A. | | Deposit date: | 2014-05-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Stabilizing the CH2 Domain of an Antibody by Engineering in an Enhanced Aromatic Sequon.
Acs Chem.Biol., 11, 2016
|
|
2BU1
 
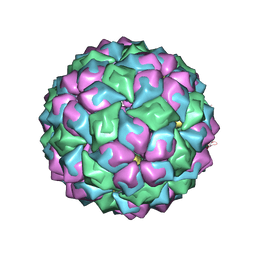 | | MS2-RNA HAIRPIN (5BRU -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *5BU*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
5J3E
 
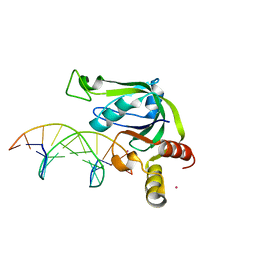 | | Crystal Structure of Human THYN1 protein in complex with 5-methylcytosine containing DNA | | Descriptor: | 5-methylcytosine containing DNA, Thymocyte nuclear protein 1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Tempel, W, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-30 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human THYN1 protein in complex with 5-methylcytosine containing DNA
To be published
|
|
4XYG
 
 | |
4XYY
 
 | |
4XYB
 
 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NADP(+) AND NaN3 | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, Formate dehydrogenase, ... | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
2BGE
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 1,2,5-THIADIAZOLIDIN-3-ONE-1,1-DIOXIDE, PROTEIN-TYROSINE PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BDM
 
 | | Structure of Cytochrome P450 2B4 with Bound Bifonazole | | Descriptor: | 1-[PHENYL-(4-PHENYLPHENYL)-METHYL]IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Zhao, Y, White, M.A, Muralidhara, B.K, Sun, L, Halpert, J.R, Stout, C.D. | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction.
J.Biol.Chem., 281, 2006
|
|
2BH8
 
 | | Combinatorial Protein 1b11 | | Descriptor: | 1B11 | | Authors: | De Bono, S, Riechmann, L, Girard, E, Williams, R.L, Winter, G. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Segment of Cold Shock Protein Directs the Folding of a Combinatorial Protein
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7SUA
 
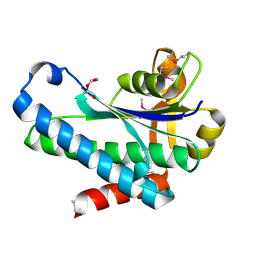 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
7BWR
 
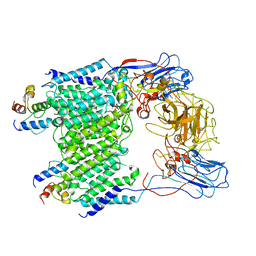 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
4QAJ
 
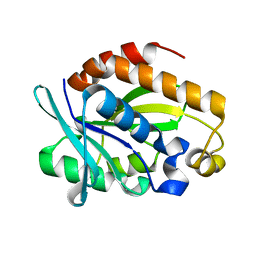 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5J98
 
 | | Crystal structure of Slow Bee Paralysis Virus at 2.6A resolution | | Descriptor: | VP1, VP2, VP3 | | Authors: | Kalynych, S, Levdansky, Y, Palkova, L, Plevka, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution.
J.Virol., 90, 2016
|
|
7T1M
 
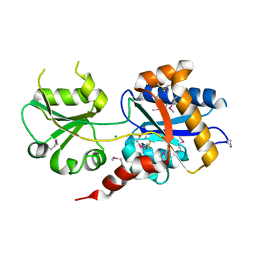 | |
4QBB
 
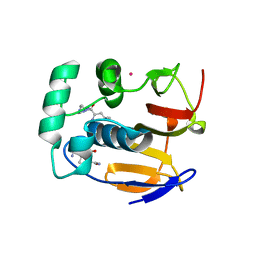 | | Structure of the foot-and-mouth disease virus leader proteinase in complex with inhibitor (N~2~-[(3S)-4-({(2R)-1-[(4-CARBAMIMIDAMIDOBUTYL)AMINO]-4-METHYL-1-OXOPENTAN-2-YL}AMINO)-3-HYDROXY-4-OXOBUTANOYL]-L-ARGINYL-L-PROLINAMIDE) | | Descriptor: | Leader protease, N~2~-[(3S)-4-({(2R)-1-[(4-carbamimidamidobutyl)amino]-4-methyl-1-oxopentan-2-yl}amino)-3-hydroxy-4-oxobutanoyl]-L-arginyl-L-prolinamide, PHOSPHATE ION, ... | | Authors: | Grishkovskaya, I, Steinberger, J, Cencic, R, Juliano, M.A, Juliano, L, Skern, T. | | Deposit date: | 2014-05-07 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Foot-and-mouth disease virus leader proteinase: Structural insights into the mechanism of intermolecular cleavage.
Virology, 468-470C, 2014
|
|
7T1S
 
 | |
2C49
 
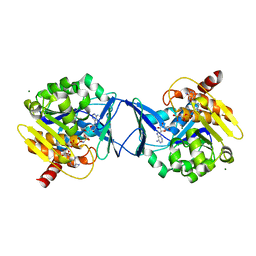 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | ADENOSINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4QCI
 
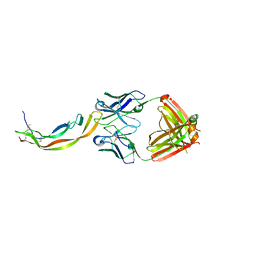 | | PDGF-B blocking antibody bound to PDGF-BB | | Descriptor: | Platelet-derived growth factor subunit B, anti-PDGF-BB antibody - Light Chain, anti-PDGF-BB antibody - Heavy chain | | Authors: | Kuai, J, Mosyak, L, Tam, M, LaVallie, E, Pullen, N, Carven, G. | | Deposit date: | 2014-05-12 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Binding Mode of Action of a Blocking Anti-Platelet-Derived Growth Factor (PDGF)-B Monoclonal Antibody, MOR8457, Reveals Conformational Flexibility and Avidity Needed for PDGF-BB To Bind PDGF Receptor-beta.
Biochemistry, 54, 2015
|
|
4XOW
 
 | |
5J3J
 
 | | Crystal Structure of human DPP-IV in complex with HL1 | | Descriptor: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
5JMQ
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP3 | | Descriptor: | 1,2-ETHANEDIOL, 9-[(5E)-7-carbamimidamido-5,6,7-trideoxy-beta-D-ribo-hept-5-enofuranosyl]-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
Febs J., 284, 2017
|
|
5J7R
 
 | | 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-06 | | Release date: | 2016-04-20 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To Be Published
|
|
