5Y9V
 
 | | Crystal structure of diamondback moth ryanodine receptor N-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor 1 | | Authors: | Lin, L, Yuchi, Z. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Crystal structure of ryanodine receptor N-terminal domain from Plutella xylostella reveals two potential species-specific insecticide-targeting sites.
Insect Biochem. Mol. Biol., 92, 2017
|
|
4IOU
 
 | |
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6AXI
 
 | | PawL-Derived Peptide PLP-2 | | Descriptor: | ASP-LEU-PHE-VAL-PRO-PRO-ILE-ASP | | Authors: | Fisher, M, Mylne, J.S, Howard, M.J. | | Deposit date: | 2017-09-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A family of small, cyclic peptides buried in preproalbumin since the Eocene epoch.
Plant Direct, 2, 2018
|
|
6JWG
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6B9K
 
 | |
4IRJ
 
 | | Structure of the mouse CD1d-4ClPhC-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Nemcovic, M, Zajonc, D.M. | | Deposit date: | 2013-01-14 | | Release date: | 2013-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection.
J.Immunol., 191, 2013
|
|
6BKL
 
 | | Influenza A M2 transmembrane domain bound to rimantadine | | Descriptor: | (1S)-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-amine, Matrix protein 2, RIMANTADINE | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters.
J. Am. Chem. Soc., 140, 2018
|
|
6CI4
 
 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis soaked with UDP-4-amino-4,6-dideoxy-L-AltNAc | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-amino-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, formyltransferase PseJ | | Authors: | Harb, I, Reimer, J.M, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.824068 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6LUL
 
 | | NMR structure and dynamics studies of yeast respiratory super-complex factor 2 in micelles | | Descriptor: | Respiratory supercomplex factor 2, mitochondrial | | Authors: | Zhou, S, Pontus, P, Peter, B, Maler, L, Adelroth, P. | | Deposit date: | 2020-01-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics Studies of Yeast Respiratory Supercomplex Factor 2.
Structure, 29, 2021
|
|
3UM0
 
 | |
7BXZ
 
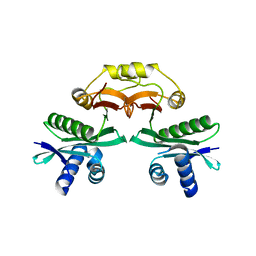 | |
4Q1B
 
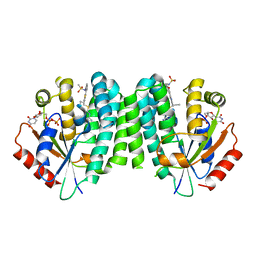 | |
4Q1D
 
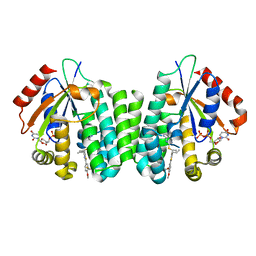 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 9 {2-{[(1R)-1-{2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-propyl-1,3-thiazol-4-yl}ethyl]sulfanyl}pyrimidine-4,6-diamine} | | Descriptor: | (R)-2-((1-(2-(3-(2-fluoroethoxy)-4-methoxyphenyl)-5-propylthiazol-4-yl)ethyl)thio)pyrimidine-4,6-diamine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
6BKK
 
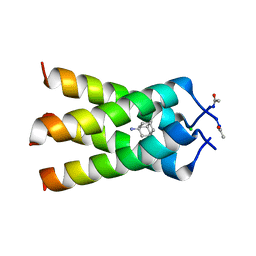 | | Influenza A M2 transmembrane domain bound to amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters.
J. Am. Chem. Soc., 140, 2018
|
|
7ZB7
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant Y54F at 1.63 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
7ZB6
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant C44S at 2.12 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
7ZB8
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant K61A at 2.48 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
3GK7
 
 | | Crystal structure of 4-hydroxybutyrate CoA-Transferase from Clostridium aminobutyricum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-Hydroxybutyrate CoA-transferase, SPERMIDINE | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M. | | Deposit date: | 2009-03-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum
Biol.Chem., 390, 2009
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
6LBH
 
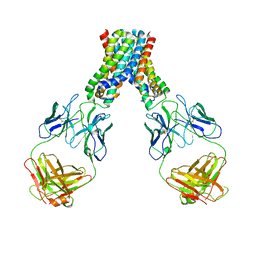 | |
5AR2
 
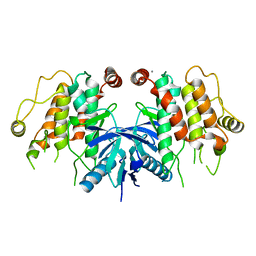 | | RIP2 Kinase Catalytic Domain (1 - 310) | | Descriptor: | CALCIUM ION, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
6BMZ
 
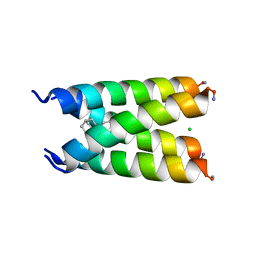 | | Influenza A M2 transmembrane domain bound to a spiroadamantane inhibitor | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, DeGrado, W.F. | | Deposit date: | 2017-11-15 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters.
J. Am. Chem. Soc., 140, 2018
|
|
4C2I
 
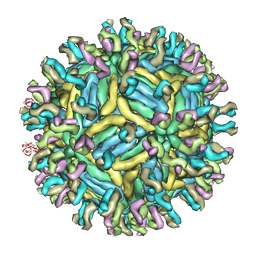 | | Cryo-EM structure of Dengue virus serotype 1 complexed with Fab fragments of human antibody 1F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fibriansah, G, Tan, J.L, de Alwis, R, Smith, S.A, Ng, T.-S, Kostyuchenko, V.A, Ibarra, K.D, Harris, E, de Silva, A, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2013-08-18 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A Potent Anti-Dengue Human Antibody Preferentially Recognizes the Conformation of E Protein Monomers Assembled on the Virus Surface.
Embo Mol.Med., 6, 2014
|
|
4LGJ
 
 | | Crystal structure and mechanism of a type III secretion protease | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
