6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
7ORF
 
 | | Crystal structure of JNK3 in complex with FMU-001-367 (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 10, ... | | Authors: | Chaikuad, A, Koch, P, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ORE
 
 | | Crystal structure of JNK3 in complex with light-activated covalent inhibitor MR-II-249 with both non-covalent and covalent binding modes (compound 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[(5Z)-9-[[4-[5-(4-fluorophenyl)-3-methyl-2-methylsulfanyl-imidazol-4-yl]pyridin-2-yl]amino]-11,12-dihydrobenzo[c][1,2]benzodiazocin-2-yl]butanamide, Mitogen-activated protein kinase 10 | | Authors: | Chaikuad, A, Reynders, M, Trauner, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OPS
 
 | | Crystal structure of haspin in complex with ZW282 (compound 2a) | | Descriptor: | 2-methylsulfanyl-10-nitro-pyrido[3,4-g]quinazoline, GLYCEROL, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synthesis and biological evaluation of Haspin inhibitors: Kinase inhibitory potency and cellular activity.
Eur.J.Med.Chem., 236, 2022
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
8QLQ
 
 | | Human MST3 (STK24) kinase in complex with macrocyclic inhibitor JA310 | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 24, macrocyclic inhibitor | | Authors: | Balourdas, D.I, Amrhein, J.A, Hanke, T, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Synthesis of Pyrazole-Based Macrocycles Leads to a Highly Selective Inhibitor for MST3.
J.Med.Chem., 67, 2024
|
|
6SY1
 
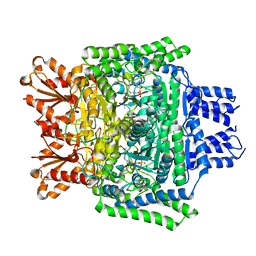 | | Crystal structure of the human 2-oxoadipate dehydrogenase DHTKD1 (E1) | | Descriptor: | MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W, Shrestha, L, Pena, I.A, Coker, J, Kolker, S, Nicola, B.B, von Delft, F, Edwards, A, Arrowsmith, C, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism.
Iucrj, 7, 2020
|
|
5O2D
 
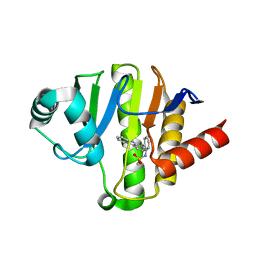 | | PARP14 Macrodomain 2 with inhibitor | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}-[2-(9~{H}-carbazol-1-yl)phenyl]methanesulfonamide | | Authors: | Uth, K, Schuller, M, Sieg, C, Wang, J, Krojer, T, Knapp, S, Riedels, K, Bracher, F, Edwards, A.M, Arrowsmith, C, Bountra, C, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
ACS Chem. Biol., 12, 2017
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6I5I
 
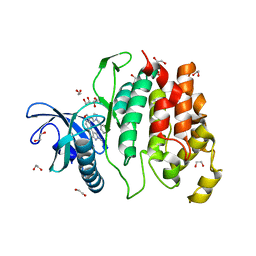 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3BHH
 
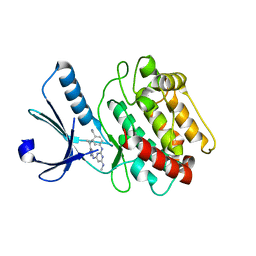 | | Crystal structure of human calcium/calmodulin-dependent protein kinase IIB isoform 1 (CAMK2B) | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II beta chain, [4-({4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-6-(methylamino)pyrimidin-2-yl}amino)phenyl]acetonitrile | | Authors: | Filippakopoulos, P, Rellos, P, Niesen, F, Burgess, N, Bullock, A, Berridge, G, Pike, A.C.W, Ugochukwu, E, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Calcium/Calmodulin-Dependent Protein Kinase IIB Isoform 1 (CAMK2B).
To be Published
|
|
8ATZ
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with SA112 (compound 2). | | Descriptor: | 2-[4-chloranyl-6-[[3-(2-phenylethoxy)phenyl]amino]pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Arifi, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8ATY
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with JP85 (compound 1). | | Descriptor: | 2-[4-chloranyl-6-(5,6,7,8-tetrahydronaphthalen-1-ylamino)pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
3C5H
 
 | | Crystal structure of the Ras homolog domain of human GRLF1 (p190RhoGAP) | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Ras homolog domain of human GRLF1 (p190RhoGAP).
To be Published
|
|
3BPU
 
 | | Crystal structure of the 3rd PDZ domain of human membrane associated guanylate kinase, C677S and C709S double mutant | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, ZINC ION | | Authors: | Pilka, E.S, Hozjan, V, Cooper, C, Pike, A.C.W, Elkins, J, Doyle, D.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the 3rd PDZ domain of human membrane associated guanylate kinase, C677S and C709S double mutant.
To be Published
|
|
3BYI
 
 | | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) | | Descriptor: | Rho GTPase activating protein 15 | | Authors: | Shrestha, L, Tickle, J, Elkins, J, Burgess-Brown, N, Johansson, C, Papagrigoriou, E, Kavanagh, K, Pike, A.C.W, Ugochukwu, E, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rho GTPase Activating Protein 15 (ARHGAP15).
To be Published
|
|
6SJM
 
 | | Crystal structure of the Retinoic Acid Receptor alpha in complex with compound 24 (JP175) | | Descriptor: | 2-[4-[3,5-bis(trifluoromethyl)phenyl]phenyl]ethanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A Novel Biphenyl-based Chemotype of Retinoid X Receptor Ligands Enables Subtype and Heterodimer Preferences.
Acs Med.Chem.Lett., 10, 2019
|
|
8C7X
 
 | | Crystal structure of BRAF in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
6I5H
 
 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6G8R
 
 | | SP140 PHD-Bromodomain complex with scFv | | Descriptor: | 1,2-ETHANEDIOL, Nuclear body protein SP140, ZINC ION, ... | | Authors: | Fairhead, M, Graslund, S, Strain-Damerell, C, Picaud, S.S, Pike, A.C.W, Pinkas, D.M, Wigren, E, Preger, C, Persson Lotsholm, H, Ossipova, E, Filippakopoulos, P, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | SP140 PHD-Bromodomain complex with scFv
To Be Published
|
|
8CPH
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
7BF6
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with remdesivir metabolite GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6I5K
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN345 (derivative of compound 12h) | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-(3-propan-2-yloxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7OAM
 
 | | Kinase domain of MERTK in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(fluoranyl)phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schroeder, M, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
