7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
1MTB
 
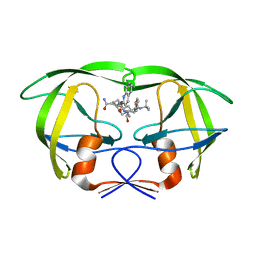 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
1MT7
 
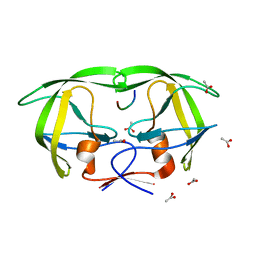 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | ACETATE ION, PROTEASE RETROPEPSIN, Substrate analogue | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
2XTZ
 
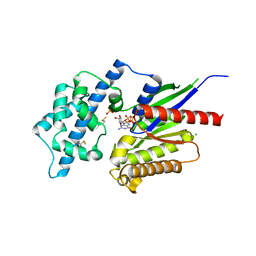 | | Crystal structure of the G alpha protein AtGPA1 from Arabidopsis thaliana | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, GUANINE NUCLEOTIDE-BINDING PROTEIN ALPHA-1 SUBUNIT, ... | | Authors: | Jones, J.C, Duffy, J.W, Machius, M, Temple, B.R.S, Dohlman, H.G, Jones, A.M. | | Deposit date: | 2010-10-13 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Crystal Structure of a Self-Activating G Protein Alpha Subunit Reveals its Distinct Mechanism of Signal Initiation
Sci.Signal., 159, 2011
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
1N49
 
 | | Viability of a Drug-Resistant HIV-1 Protease Variant: Structural Insights for Better Anti-Viral Therapy | | Descriptor: | Protease, RITONAVIR | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viability of a Drug-Resistant Human Immunodeficiency Virus Type 1 Protease Variant:
Structural Insights for Better Antiviral Therapy
J.VIROL., 77, 2003
|
|
7F4A
 
 | |
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7VS4
 
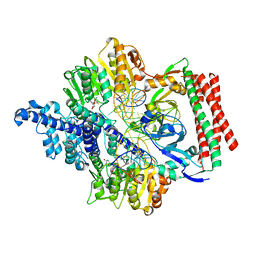 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7VRU
 
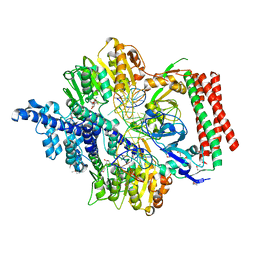 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
8JP0
 
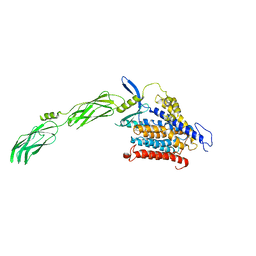 | | structure of human sodium-calciumexchanger NCX1 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, Sodium/calcium exchanger 1 | | Authors: | Dong, Y, Zhao, Y. | | Deposit date: | 2023-06-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the allosteric inhibition of human sodium-calcium exchanger NCX1 by XIP and SEA0400.
Embo J., 43, 2024
|
|
5HXW
 
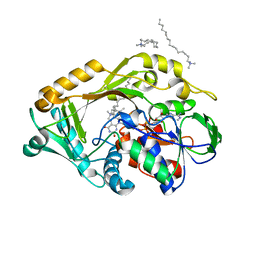 | | L-amino acid deaminase from Proteus vulgaris | | Descriptor: | CETYL-TRIMETHYL-AMMONIUM, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-01-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
5I39
 
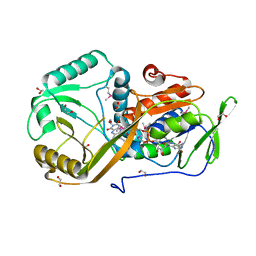 | | High resolution structure of L-amino acid deaminase from Proteus vulgaris with the deletion of the specific insertion sequence | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
3RDY
 
 | |
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
3RE1
 
 | |
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
3RDZ
 
 | |
3S69
 
 | | Crystal structure of saxthrombin | | Descriptor: | CALCIUM ION, Thrombin-like enzyme defibrase | | Authors: | Huang, K, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of saxthrombin, a thrombin-like enzyme from Gloydius saxatilis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3S9V
 
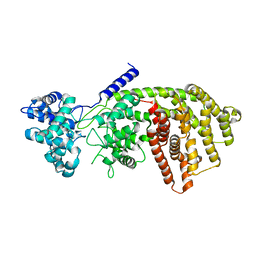 | | abietadiene synthase from Abies grandis | | Descriptor: | Abietadiene synthase, chloroplastic | | Authors: | Zhou, K, Hoy, J.A, Mann, F.M, Honzatko, R.B, Peters, R.J. | | Deposit date: | 2011-06-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis.
J.Biol.Chem., 287, 2012
|
|
7BWR
 
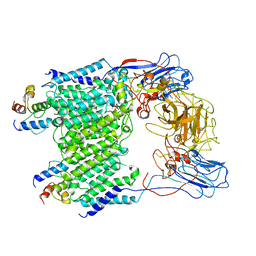 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
5C8S
 
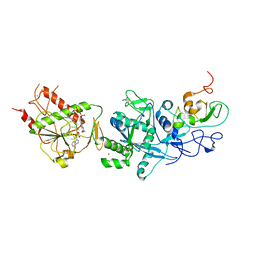 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Guanine-N7 methyltransferase, MAGNESIUM ION, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
