8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
7LUU
 
 | | Kinetic and Structural Characterization of the First B3 Metallo-beta-Lactamase with an Active Site Glutamic Acid | | Descriptor: | GLYCEROL, SULFATE ION, Subclass B3 metallo-beta-lactamase, ... | | Authors: | Wilson, L, Knaven, E, Morris, M.T, Schenk, G. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Kinetic and Structural Characterization of the First B3 Metallo-beta-Lactamase with an Active-Site Glutamic Acid.
Antimicrob.Agents Chemother., 65, 2021
|
|
2INY
 
 | |
8DTN
 
 | | The complex of nanobody 6101 with BCL11A ZF6 | | Descriptor: | B-cell lymphoma/leukemia 11A, MAGNESIUM ION, Nanobody 6101, ... | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3ZI5
 
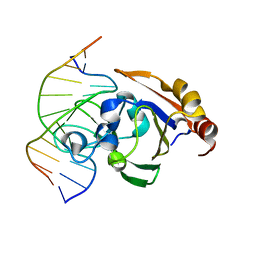 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | Descriptor: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | Authors: | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2013-01-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
8DTU
 
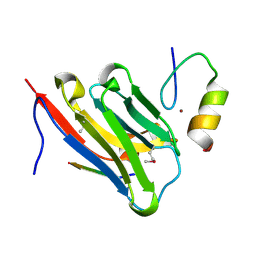 | | The complex of nanobody 5344N74D with BCL11A ZF6. | | Descriptor: | B-cell lymphoma/leukemia 11A, Nanobody 5344N74D, ZINC ION | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6Q1Q
 
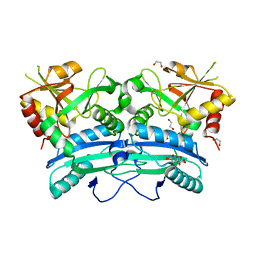 | |
6Q2T
 
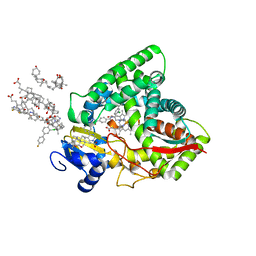 | | Human sterol 14a-demethylase (CYP51) in complex with the functionally irreversible inhibitor (R)-N-(1-(3-chloro-4'-fluoro-[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Lanosterol 14-alpha demethylase, N-[(1R)-1-(3-chloro-4'-fluoro[1,1'-biphenyl]-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[3-fluoro-5-(5-fluoropyrimidin-4-yl)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, ... | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Validation of Human Sterol 14 alpha-Demethylase (CYP51) Druggability: Structure-Guided Design, Synthesis, and Evaluation of Stoichiometric, Functionally Irreversible Inhibitors.
J.Med.Chem., 62, 2019
|
|
8DPX
 
 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
6PW7
 
 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | Descriptor: | CALCIUM ION, Stromal interaction molecule 1 | | Authors: | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-02-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
7X6S
 
 | |
7X6V
 
 | |
8E29
 
 | |
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
8E27
 
 | | RNA-free Human Dis3L2 | | Descriptor: | DIS3-like exonuclease 2 | | Authors: | Meze, K, Thomas, D.R, Joshua-Tor, L. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A shape-shifting nuclease unravels structured RNA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E28
 
 | |
8E2A
 
 | |
5Z34
 
 | |
8E7F
 
 | | Crystal structure of the autotransporter Ssp from Serratia marcescens. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Extracellular serine protease, ... | | Authors: | Hor, L, Pilapitiya, A, Panjikar, S, Paxman, J.J, Heras, B. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function.
Nat Commun, 14, 2023
|
|
3ZKF
 
 | | Structure of LC8 in complex with Nek9 phosphopeptide | | Descriptor: | DYNEIN LIGHT CHAIN 1, CYTOPLASMIC, NEK9 PROTEIN | | Authors: | Gallego, P, Velazquez-Campoy, A, Regue, L, Roig, J, Reverter, D. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Regulation of the Dynll/Lc8 Binding to Nek9 by Phosphorylation
J.Biol.Chem., 288, 2013
|
|
3ZNI
 
 | | Structure of phosphoTyr363-Cbl-b - UbcH5B-Ub - ZAP-70 peptide complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL-B, ... | | Authors: | Dou, H, Buetow, L, Sibbet, G.J, Cameron, K, Huang, D.T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Essentiality of a Non-Ring Element in Priming Donor Ubiquitin for Catalysis by a Monomeric E3.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6PXS
 
 | | Crystal structure of iminodiacetate oxidase (IdaA) from Chelativorans sp. BNC1 | | Descriptor: | FAD dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Jun, S.Y, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structural and biochemical characterization of iminodiacetate oxidase from Chelativorans sp. BNC1.
Mol.Microbiol., 112, 2019
|
|
3ZC4
 
 | | The structure of Csa5 from Sulfolobus solfataricus. | | Descriptor: | DI(HYDROXYETHYL)ETHER, SSO1398 | | Authors: | Reeks, J, Anderson, L, White, M.F, Naismith, J.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the Archaeal Cascade Subunit Csa5: Relating the Small Subunits of Crispr Effector Complexes.
RNA Biol., 10, 2013
|
|
6PUQ
 
 | |
