7LO8
 
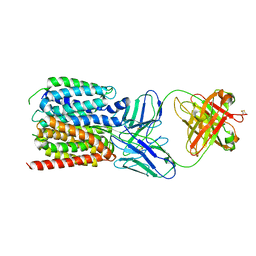 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO7
 
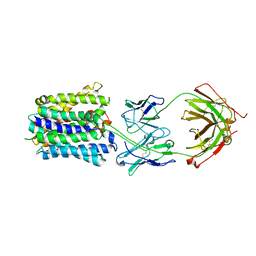 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
8DT0
 
 | |
7QUI
 
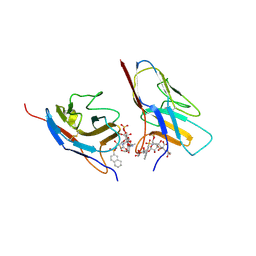 | | Crystal structure of the N-terminal domain of Siglec-8 in complex with sulfonamide sialoside analogue | | Descriptor: | (2~{S},4~{S},5~{R},6~{R})-5-acetamido-2-[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-acetamido-2-(hydroxymethyl)-4,6-bis(oxidanyl)oxan-3-yl]oxy-3,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-4-yl]oxy-6-[(1~{R},2~{R})-3-(naphthalen-2-ylsulfonylamino)-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Oyenarte, I, Atxabal, U, Nycholat, C, Franconetti, A, Quintana, J.I, Delgado, S, Unione, L, Paulson, J, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
8TQ1
 
 | | HIV-1 BG505 Env SOSIP in complex with bovine Fab Bess4 and non-human primate Fab RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bovine Bess4 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
4MOB
 
 | | Acyl-Coenzyme A thioesterase 12 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Acyl-coenzyme A thioesterase 12, COENZYME A | | Authors: | Swarbrick, C.M.D, Forwood, J.K. | | Deposit date: | 2013-09-12 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for regulation of the human acetyl-CoA thioesterase 12 and interactions with the steroidogenic acute regulatory protein-related lipid transfer (START) domain.
J.Biol.Chem., 289, 2014
|
|
4MOC
 
 | | Human Acyl-coenzyme A Thioesterase 12 | | Descriptor: | Acyl-coenzyme A thioesterase 12, COENZYME A | | Authors: | Swarbrick, C.M.D, Forwood, J.K. | | Deposit date: | 2013-09-12 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for regulation of the human acetyl-CoA thioesterase 12 and interactions with the steroidogenic acute regulatory protein-related lipid transfer (START) domain.
J.Biol.Chem., 289, 2014
|
|
9EOU
 
 | |
6ICI
 
 | | Crystal structure of human MICAL3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, [F-actin]-monooxygenase MICAL3 | | Authors: | Hwang, K.Y, Kim, J.S. | | Deposit date: | 2018-09-06 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic insights into flavin-containing monooxygenase and calponin-homology domains in human MICAL3.
Iucrj, 7, 2020
|
|
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
5X68
 
 | | Crystal Structure of Human KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6P
 
 | | Crystal structure of Pseudomonas fluorescens KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6Q
 
 | | Crystal structure of Pseudomonas fluorescens KMO in complex with Ro 61-8048 | | Descriptor: | 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6R
 
 | |
7MCF
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-6-fluoro-5-[(S)-(3-fluoropyridin-2-yl)(oxan-4-yl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A search for structural diversity.
Bioorg.Med.Chem.Lett., 44, 2021
|
|
1HHI
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), INFLUENZA A MATRIX PROTEIN M1 (RESIDUES 58-66) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
8INV
 
 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
6YSW
 
 | | E. coli anaerobic trifunctional enzyme subunit-alpha in complex with coenzyme A | | Descriptor: | COENZYME A, Fatty acid oxidation complex subunit alpha, SULFATE ION | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2020-04-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes
Structure, 2023
|
|
6YSV
 
 | |
6IQ6
 
 | | Crystal structure of GAPDH | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Park, J.B, Park, H.Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Study of Monomethyl Fumarate-Bound Human GAPDH.
Mol.Cells, 42, 2019
|
|
9CSY
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to PF-07957472 | | Descriptor: | 2-methyl-5-(4-methylpiperazin-1-yl)-N-{1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]cyclopropyl}benzamide, Papain-like protease, ZINC ION, ... | | Authors: | Mashalidis, E.H, Chang, J.S, Wu, H, Garnsey, M. | | Deposit date: | 2024-07-24 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Discovery of SARS-CoV-2 papain-like protease (PL pro ) inhibitors with efficacy in a murine infection model.
Sci Adv, 10, 2024
|
|
9EYU
 
 | | Human PRMT5 in complex with AZ compound 1 | | Descriptor: | (1~{S})-2-[(2-carbamimidamido-1,3-thiazol-5-yl)methyl]-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9EYW
 
 | | Human PRMT5 in complex with AZ compound 21 | | Descriptor: | (3~{S})-2-[(5-azanyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9EYX
 
 | | Human PRMT5 in complex with AZ compound 28 | | Descriptor: | (3~{S})-2-[(5-azanyl-6-fluoranyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9EYV
 
 | | Human PRMT5 in complex with AZ compound 12 | | Descriptor: | (1~{S})-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-2-(1~{H}-pyrrolo[3,2-b]pyridin-2-ylmethyl)-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
