317D
 
 | |
327D
 
 | |
393D
 
 | |
322D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
394D
 
 | |
321D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
332D
 
 | |
318D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCC(BR)5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(CBR)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
358D
 
 | |
320D
 
 | | CRYSTAL STRUCTURES OF D(CCGGGCCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3'), SPERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
1FLV
 
 | |
396D
 
 | |
319D
 
 | | CRYSTAL STRUCTURES OF D(CCGGG(BR)5CCCGG)-ORTHOGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*(CBR)P*CP*CP*GP*G)-3'), THERMINE | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
438D
 
 | |
3Q91
 
 | | Crystal Structure of Human Uridine Diphosphate Glucose Pyrophosphatase (NUDT14) | | Descriptor: | Uridine diphosphate glucose pyrophosphatase | | Authors: | Tresaugues, L, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Uridine Diphosphate Glucose Pyrophosphatase (NUDT14)
To be Published
|
|
4BCT
 
 | | Crystal structure of kiwi-fruit allergen Act d 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, THAUMATIN-LIKE PROTEIN | | Authors: | Pavkov-Keller, T, Bublin, M, Jankovic, M, Breiteneder, H, Keller, W. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal Structure of Kiwi-Fruit Allergen Act D 2
To be Published
|
|
4BUH
 
 | | Human IgE against the major allergen Bet v 1 - Crystal structure of clone M0418 scFv | | Descriptor: | 1,2-ETHANEDIOL, CLONE M0418 SCFV, DI(HYDROXYETHYL)ETHER | | Authors: | Davies, A.M, Levin, M, Lilljekvist, M, Carlsson, F, Gould, H.J, Sutton, B.J, Ohlin, M. | | Deposit date: | 2013-06-20 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Ige Against the Major Allergen Bet V 1 - Defining an Epitope with Limited Cross-Reactivity between Different Pr-10 Family Proteins
Clin.Exp.Allergy, 44, 2014
|
|
4N4Z
 
 | | Trypanosoma brucei procathepsin B structure solved by Serial Microcrystallography using synchrotron radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gati, C, Bourenkov, G, Klinge, M, Rehders, D, Stellato, F, Oberthuer, D, White, T.A, Yevanov, O, Sommer, B.P, Mogk, S, Duszenko, M, Betzel, C, Schneider, T.R, Chapman, H.N, Redecke, L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Serial crystallography on in vivo grown microcrystals using synchrotron radiation.
IUCrJ, 1, 2014
|
|
3ZPR
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-methyl-2-(piperazin-1-yl) quinoline bound | | Descriptor: | 4-METHYL-2-(PIPERAZIN-1-YL) QUINOLINE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
3PH9
 
 | | Crystal structure of the human anterior gradient protein 3 | | Descriptor: | Anterior gradient protein 3 homolog | | Authors: | Nguyen, V.D, Ruddock, L.W, Salin, M, Wierenga, R.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of human anterior gradient protein 3.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
4GZK
 
 | |
4CY7
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form II | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
4BJ0
 
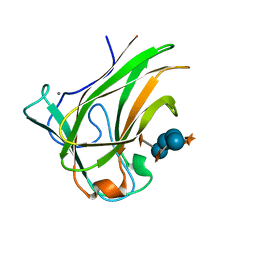 | | Xyloglucan binding module (CBM4-2 X2-L110F) in complex with branched xyloses | | Descriptor: | CALCIUM ION, XYLANASE, alpha-D-glucopyranose, ... | | Authors: | Schantz, L, Hakansson, M, Logan, D.T, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2013-04-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Carbohydrate Binding Module Recognition of Xyloglucan Defined by Polar Contacts with Branching Xyloses and Ch-Pi Interactions.
Proteins, 82, 2014
|
|
3ZPQ
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-(piperazin-1- yl)-1H-indole bound (compound 19) | | Descriptor: | 4-(PIPERAZIN-1-YL)-1H-INDOLE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
4CML
 
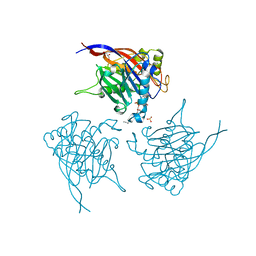 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
