1DS1
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DRT
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND PROCLAVAMINIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, 5-AMINO-3-HYDROXY-2-(2-OXO-AZETIDIN-1-YL)-PENTANOIC ACID, CLAVAMINATE SYNTHASE 1, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DS0
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE | | Descriptor: | ACETATE ION, CLAVAMINATE SYNTHASE 1, SULFATE ION | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
7VJL
 
 | | The crystal structure of FGFR4 kinase domain in complex with N-(5-cyano-4-((2-methoxyethyl)amino)pyridin-2-yl)-7-(2,2,2-trifluoroacetyl)-3,4-dihydro-1,8-naphthyridine-1(2H)-carboxamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-(aminomethyl)-4-(2-methoxyethylamino)pyridin-2-yl]-7-[2,2,2-tris(fluoranyl)ethanoyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.900173 Å) | | Cite: | Characterization of an aromatic trifluoromethyl ketone as a new warhead for covalently reversible kinase inhibitor design.
Bioorg.Med.Chem., 50, 2021
|
|
4H5N
 
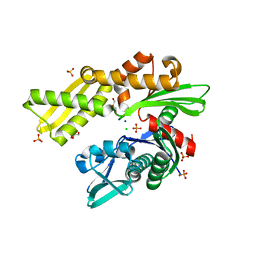 | | HSC70 NBD with PO4, Na, Cl | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
4H5W
 
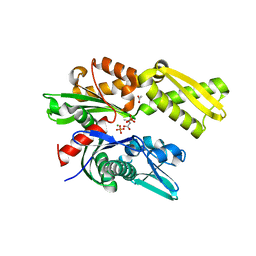 | | HSC70 NBD with betaine | | Descriptor: | ACETATE ION, Heat shock cognate 71 kDa protein, MAGNESIUM ION, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
7M4X
 
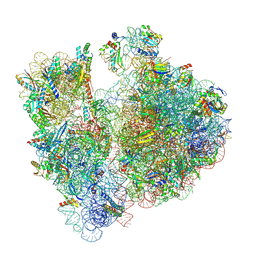 | | A. baumannii Ribosome-Eravacycline complex: P-site tRNA 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4U
 
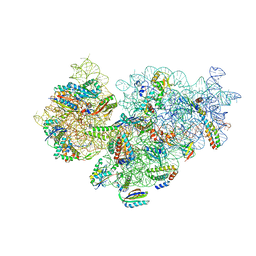 | | A. baumannii Ribosome-Eravacycline complex: 30S | | Descriptor: | 16s Ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4Y
 
 | | A. baumannii Ribosome-Eravacycline complex: E-site tRNA 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
4H5T
 
 | | HSC70 NBD with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock cognate 71 kDa protein, MAGNESIUM ION, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
7M4Z
 
 | | A. baumannii Ribosome-Eravacycline complex: hpf-bound 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4V
 
 | | A. baumannii Ribosome-Eravacycline complex: 50S | | Descriptor: | 23s ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4W
 
 | | A. baumannii Ribosome-Eravacycline complex: Empty 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
4H5V
 
 | | HSC70 NBD with Mg | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
4H5R
 
 | | HSC70 NBD with Na, Cl and glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | Authors: | Stec, B. | | Deposit date: | 2012-09-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition.
Biochimie, 108, 2015
|
|
6KMH
 
 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
8J9N
 
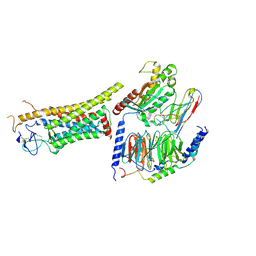 | | Gq bound FZD1 in ligand-free state | | Descriptor: | Frizzled-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8J9O
 
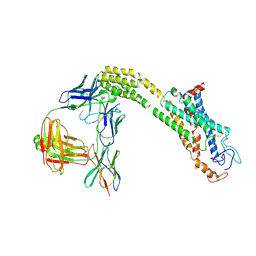 | | Cryo-EM structure of inactive FZD1 | | Descriptor: | Frizzled-1,Soluble cytochrome b562, anti-BRIL Fab Heavy Chain, anti-BRIL Fab Light Chain, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
3KE1
 
 | |
2JX0
 
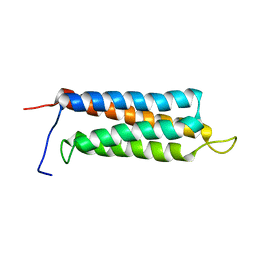 | | The paxillin-binding domain (PBD) of G Protein Coupled Receptor (GPCR)-kinase (GRK) interacting protein 1 (GIT1) | | Descriptor: | ARF GTPase-activating protein GIT1 | | Authors: | Zhang, Z, Guibao, C.D, Simmerman, J.A, Zheng, J. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs.
J.Biol.Chem., 283, 2008
|
|
3Q8H
 
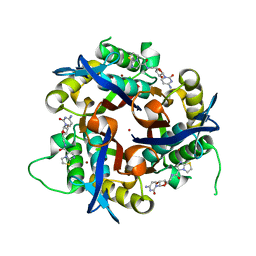 | | Crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from burkholderia pseudomallei in complex with cytidine derivative EBSI01028 | | Descriptor: | 1,2-ETHANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-deoxy-5'-[(imidazo[2,1-b][1,3]thiazol-5-ylcarbonyl)amino]cytidine, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cytidine derivatives as IspF inhibitors of Burkolderia pseudomallei.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ONA
 
 | |
4M84
 
 | |
4LWT
 
 | | The 1.6A Crystal Structure of Humanized Xenopus MDM2 with RO5027344 | | Descriptor: | (3S)-3-[(3R)-1-acetylpiperidin-3-yl]-6-chloro-3-(3-chlorobenzyl)-1,3-dihydro-2H-indol-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Kammlott, U. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
4LWU
 
 | | The 1.14A Crystal Structure of Humanized Xenopus MDM2 with RO5499252 | | Descriptor: | (2'S,3R,4'S,5'R)-N-(4-carbamoylphenyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
