2ZW0
 
 | | Crystal structure of a Streptococcal protein G B1 mutant | | Descriptor: | Protein LG, SULFATE ION | | Authors: | Watanabe, H, Matsumaru, H, Odahara, T, Suto, K, Honda, S. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimizing pH response of affinity between protein G and IgG Fc: how electrostatic modulations affect protein-protein interactions.
J.Biol.Chem., 284, 2009
|
|
2ZW1
 
 | | Crystal structure of a Streptococcal protein G B1 mutant | | Descriptor: | Protein LG | | Authors: | Watanabe, H, Matsumaru, H, Odahara, T, Suto, K, Honda, S. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimizing pH response of affinity between protein G and IgG Fc: how electrostatic modulations affect protein-protein interactions.
J.Biol.Chem., 284, 2009
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
6JOM
 
 | | Crystal structure of lipoate protein ligase from Mycoplasma hyopneumoniae | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, Lipoate--protein ligase | | Authors: | Zhang, H, Chen, H, Ma, G. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Identification and Structural Analysis of a New Lipoate Protein Ligase inMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 10, 2020
|
|
6K1T
 
 | |
6LFB
 
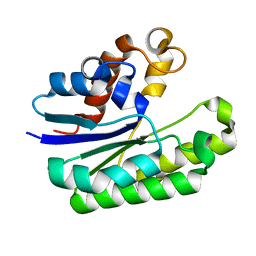 | | E. coli Thioesterase I mutant DG | | Descriptor: | Acyl-CoA thioesterase I also functions as protease I | | Authors: | Deng, X, Chen, L, Yang, G. | | Deposit date: | 2019-12-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-guided reshaping of the acyl binding pocket of 'TesA thioesterase enhances octanoic acid production in E. coli.
Metab. Eng., 61, 2020
|
|
3NCO
 
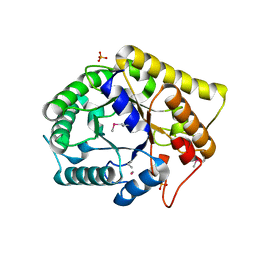 | | Crystal structure of FnCel5A from F. nodosum Rt17-B1 | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, peptide (ALA)(ASN)(GLU), ... | | Authors: | Zheng, B.S, Yang, W, Wang, Y, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2010-06-05 | | Release date: | 2011-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of FnCel5A from F. nodosum Rt17-B1
To be Published
|
|
8WZB
 
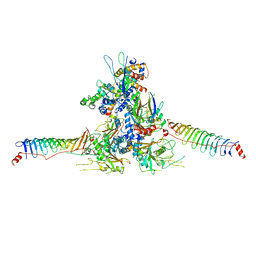 | | RS head-neck monomer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
8X2U
 
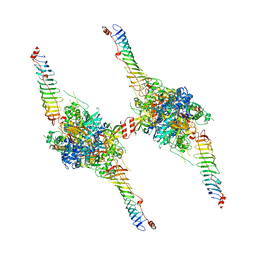 | | Radial spoke head-neck dimer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
4LI6
 
 | | TANKYRASE-1 Complexed with small molecule inhibitor N-[(4-oxo-3,4-dihydroquinazolin-2-yl)methyl]-3-phenyl-N-(thiophen-2-ylmethyl)propanamide | | Descriptor: | N-[(4-oxo-3,4-dihydroquinazolin-2-yl)methyl]-3-phenyl-N-(thiophen-2-ylmethyl)propanamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
4LI8
 
 | | TANKYRASE-1 complexed with small molecule inhibitor 2-[4-(4-fluorobenzoyl)piperidin-1-yl]-N-[(4-oxo-3,5,7,8-tetrahydro-4H-pyrano[4,3-d]pyrimidin-2-yl)methyl]-N-(thiophen-2-ylmethyl)acetamide | | Descriptor: | 2-[4-(4-fluorobenzoyl)piperidin-1-yl]-N-[(4-oxo-3,5,7,8-tetrahydro-4H-pyrano[4,3-d]pyrimidin-2-yl)methyl]-N-(thiophen-2-ylmethyl)acetamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
4LI7
 
 | | TANKYRASE-1 complexed with small molecule inhibitor 4-chloro-5-cyano-N-{2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl}-2-methoxybenzamide | | Descriptor: | 4-chloro-5-cyano-N-{2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl}-2-methoxybenzamide, SODIUM ION, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
4M7I
 
 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
4M5D
 
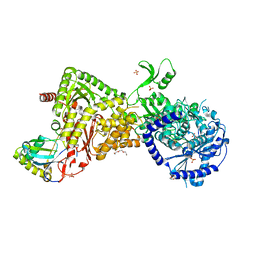 | |
5EWI
 
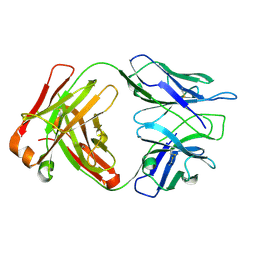 | |
5EKU
 
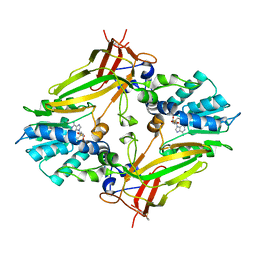 | |
7FIA
 
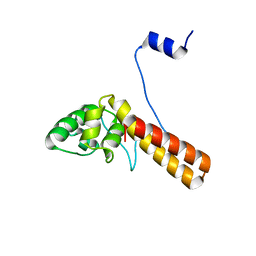 | | Structure of AcrIF23 | | Descriptor: | AcrIF23 | | Authors: | Ren, J, Yue, F. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23.
J.Biol.Chem., 298, 2022
|
|
7CNL
 
 | | Crystal structure of TEAD3 in complex with VT105 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, N-[(1S)-1-pyridin-2-ylethyl]-8-[4-(trifluoromethyl)phenyl]quinoline-3-carboxamide, ... | | Authors: | Tang, T.T, Konradi, A.W. | | Deposit date: | 2020-08-01 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small Molecule Inhibitors of TEAD Auto-palmitoylation Selectively Inhibit Proliferation and Tumor Growth of NF2 -deficient Mesothelioma.
Mol.Cancer Ther., 20, 2021
|
|
7E7Y
 
 | |
7E7X
 
 | |
7E88
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
8HN6
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
