4U45
 
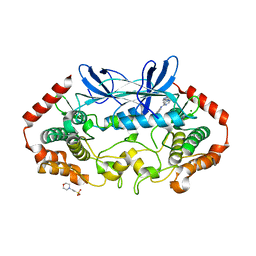 | | MAP4K4 in complex with inhibitor (compound 25) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(1H-pyrazol-4-yl)-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8HBT
 
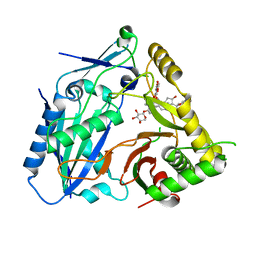 | | AmAT7-3 mutant A310G | | Descriptor: | AmAT7-3-A310G, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-31 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
8H8I
 
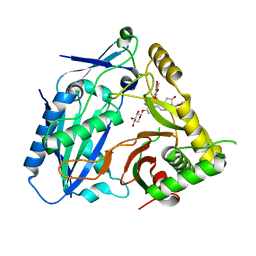 | | Triterpenoid saponin acetyltransferase, AmAT7-3 | | Descriptor: | AmAT7-3, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
4U44
 
 | | MAP4K4 in complex with inhibitor (compound 16) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-phenyl-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6EGE
 
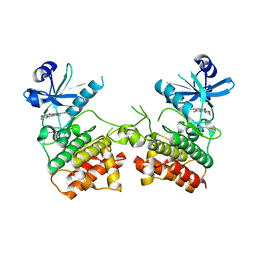 | |
7LRT
 
 | |
7LRS
 
 | |
7CBZ
 
 | | Crystal structure of T2R-TTL-A31 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-[2-[4-(2-cyclopropylethanoyl)piperazin-1-yl]ethoxy]phenyl]pyridin-2-yl]-N-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
J.Med.Chem., 64, 2021
|
|
7YF1
 
 | |
3DBZ
 
 | | human surfactant protein D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Head, J.F. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of recombinant surfactant protein D with lipopolysaccharide: conformation and orientation of bound protein by IRRAS and simulations.
Biochemistry, 47, 2008
|
|
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | Descriptor: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6.8 Å) | | Cite: | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7TYJ
 
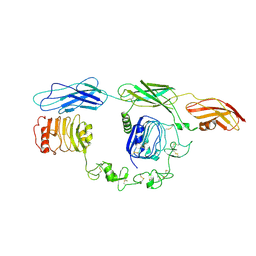 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was focused on one of two halves with C1 symmetry applied | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYK
 
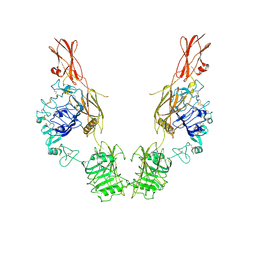 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
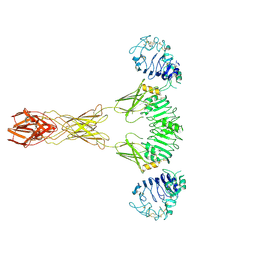 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4J6O
 
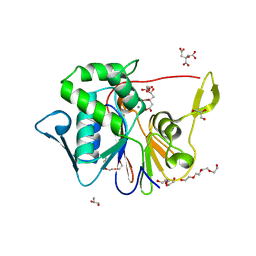 | | Crystal Structure of the Phosphatase Domain of C. thermocellum (Bacterial) PnkP | | Descriptor: | CITRIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wang, L, Smith, P, Shuman, S. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the 2',3' phosphatase component of the bacterial Pnkp-Hen1 RNA repair system.
Nucleic Acids Res., 41, 2013
|
|
7M3I
 
 | |
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
7X8B
 
 | |
7X8G
 
 | |
7X8F
 
 | |
7X88
 
 | |
6N0Q
 
 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
5VAL
 
 | | BRAF in Complex with N-(3-(tert-butyl)phenyl)-4-methyl-3-(6-morpholinopyrimidin-4-yl)benzamide | | Descriptor: | N-(3-tert-butylphenyl)-4-methyl-3-[6-(morpholin-4-yl)pyrimidin-4-yl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
