4YBM
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (7b) | | Descriptor: | GLYCEROL, N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-3,4-dimethoxybenzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBS
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7g) | | Descriptor: | DIMETHYL SULFOXIDE, N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAX
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]benzenesulfonamide (5g) | | Descriptor: | GLYCEROL, N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, SULFATE ION, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-methoxybenzene-1-sulfonamide (5b) | | Descriptor: | N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-4-methoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ZINC ION | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l) | | Descriptor: | DIMETHYL SULFOXIDE, N-(1,3-dimethyl-2-oxo-6-{3-[(3S)-tetrahydrofuran-3-ylmethoxy]phenoxy}-2,3-dihydro-1H-benzimidazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YC9
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (8i) | | Descriptor: | GLYCEROL, N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-3,4-dimethoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4XEB
 
 | | The structure of P. funicolosum Cel7A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2014-12-23 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering enhanced cellobiohydrolase activity
Nat Commun, 9(1), 2018
|
|
4ZBN
 
 | |
1DPI
 
 | |
1MUP
 
 | | PHEROMONE BINDING TO TWO RODENT URINARY PROTEINS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN | | Authors: | Bocskei, Z, Flower, D.R, Groom, C.R, Phillips, S.E.V, North, A.C.T. | | Deposit date: | 1992-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography.
Nature, 360, 1992
|
|
2K0N
 
 | |
2IXA
 
 | | A-zyme, N-acetylgalactosaminidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
2IXB
 
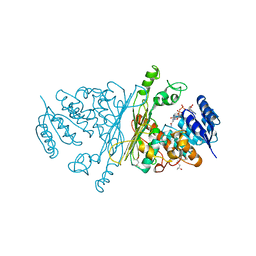 | | Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
2HHD
 
 | | OXYGEN AFFINITY MODULATION BY THE N-TERMINI OF THE BETA-CHAINS IN HUMAN AND BOVINE HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) (ALPHA CHAIN), HEMOGLOBIN (DEOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gilliland, G.L, Pechik, I, Fronticelli, C, Ji, X. | | Deposit date: | 1994-09-29 | | Release date: | 1995-01-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic, molecular modeling, and biophysical characterization of the valine beta 67 (E11)-->threonine variant of hemoglobin.
Biochemistry, 35, 1996
|
|
2HHE
 
 | | OXYGEN AFFINITY MODULATION BY THE N-TERMINI OF THE BETA CHAINS IN HUMAN AND BOVINE HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) (ALPHA CHAIN), HEMOGLOBIN (DEOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gilliland, G.L, Pechik, I, Fronticelli, C, Ji, X. | | Deposit date: | 1994-09-29 | | Release date: | 1995-01-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloride ion independence of the Bohr effect in a mutant human hemoglobin beta (V1M+H2deleted).
J.Biol.Chem., 269, 1994
|
|
2KLV
 
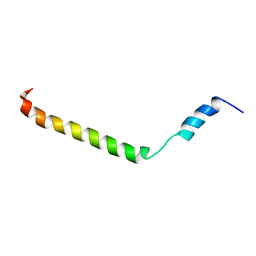 | | Membrane-bound structure of the Pf1 major coat protein in DHPC micelle | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Son, W, Mukhopadhyay, R, Valafar, H, Opella, S.J. | | Deposit date: | 2009-07-08 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage-induced alignment of membrane proteins enables the measurement and structural analysis of residual dipolar couplings with dipolar waves and lambda-maps.
J.Am.Chem.Soc., 131, 2009
|
|
2KPH
 
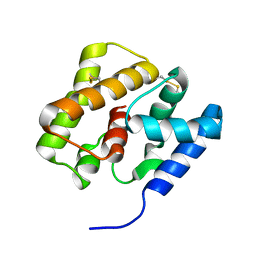 | | NMR Structure of AtraPBP1 at pH 4.5 | | Descriptor: | Pheromone binding protein | | Authors: | Ames, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Navel Orangeworm Moth Pheromone-Binding Protein (AtraPBP1): Implications for pH-Sensitive Pheromone Detection .
Biochemistry, 49, 2010
|
|
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
2LJ2
 
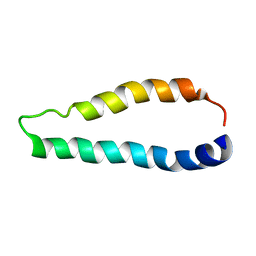 | | Integral membrane core domain of the mercury transporter MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Das, B.B, Nothnagel, H.J, Lu, G.J, Son, W, Park, S, Tian, Y.B, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-09-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of a membrane protein in proteoliposomes.
J.Am.Chem.Soc., 134, 2012
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
5US4
 
 | | Crystal structure of human KRAS G12D mutant in complex with GDP | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T, Kaplan, A, Stockwell, B.R, Tong, L. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Multivalent Small-Molecule Pan-RAS Inhibitors.
Cell, 168, 2017
|
|
5UWZ
 
 | |
5USJ
 
 | | Crystal Structure of human KRAS G12D mutant in complex with GDPNP | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Huang, C.S, Kaplan, A, Stockwell, B.R, Tong, L. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Multivalent Small-Molecule Pan-RAS Inhibitors.
Cell, 168, 2017
|
|
5UQW
 
 | | Crystal structure of human KRAS G12V mutant in complex with GDP | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Huang, C.S, Kaplan, A, Stockwell, B.R, Tong, L. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multivalent Small-Molecule Pan-RAS Inhibitors.
Cell, 168, 2017
|
|
5UQ9
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase with ((4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl)methyl dihydrogen phosphate | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, [(4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl dihydrogen phosphate | | Authors: | Leonard, P.G. | | Deposit date: | 2017-02-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation.
Cell Rep, 26, 2019
|
|
