4LI7
 
 | | TANKYRASE-1 complexed with small molecule inhibitor 4-chloro-5-cyano-N-{2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl}-2-methoxybenzamide | | Descriptor: | 4-chloro-5-cyano-N-{2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl}-2-methoxybenzamide, SODIUM ION, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
4LI6
 
 | | TANKYRASE-1 Complexed with small molecule inhibitor N-[(4-oxo-3,4-dihydroquinazolin-2-yl)methyl]-3-phenyl-N-(thiophen-2-ylmethyl)propanamide | | Descriptor: | N-[(4-oxo-3,4-dihydroquinazolin-2-yl)methyl]-3-phenyl-N-(thiophen-2-ylmethyl)propanamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
4KRS
 
 | | Tankyrase-1 complexed with small molecule inhibitor | | Descriptor: | 4-tert-butyl-N-(5,6-dihydro[1,3]thiazolo[2,3-c][1,2,4]triazol-3-yl)benzamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Stams, T, Kirby, C. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-Efficiency Relationship of [1,2,4]Triazol-3-ylamines as Novel Nicotinamide Isosteres that Inhibit Tankyrases.
J.Med.Chem., 56, 2013
|
|
4LI8
 
 | | TANKYRASE-1 complexed with small molecule inhibitor 2-[4-(4-fluorobenzoyl)piperidin-1-yl]-N-[(4-oxo-3,5,7,8-tetrahydro-4H-pyrano[4,3-d]pyrimidin-2-yl)methyl]-N-(thiophen-2-ylmethyl)acetamide | | Descriptor: | 2-[4-(4-fluorobenzoyl)piperidin-1-yl]-N-[(4-oxo-3,5,7,8-tetrahydro-4H-pyrano[4,3-d]pyrimidin-2-yl)methyl]-N-(thiophen-2-ylmethyl)acetamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
5EWI
 
 | |
3UDC
 
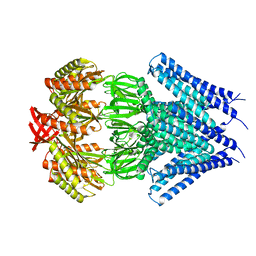 | | Crystal structure of a membrane protein | | Descriptor: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | Authors: | Li, W, Ge, J, Yang, M. | | Deposit date: | 2011-10-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.355 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4M7I
 
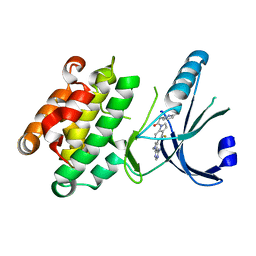 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
4M5D
 
 | |
3NCO
 
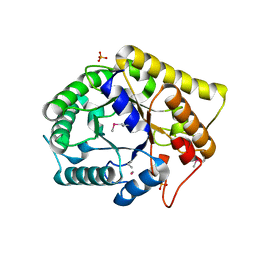 | | Crystal structure of FnCel5A from F. nodosum Rt17-B1 | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, peptide (ALA)(ASN)(GLU), ... | | Authors: | Zheng, B.S, Yang, W, Wang, Y, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2010-06-05 | | Release date: | 2011-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of FnCel5A from F. nodosum Rt17-B1
To be Published
|
|
8HIH
 
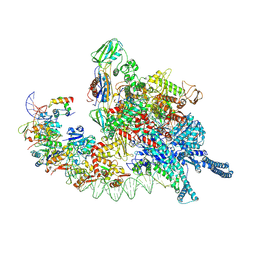 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HML
 
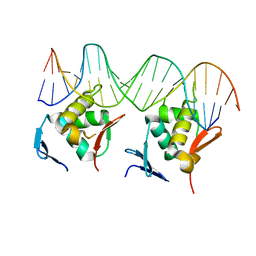 | |
6LBP
 
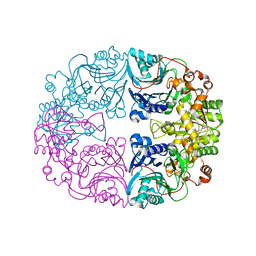 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | Descriptor: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | Authors: | Yi, Z, Cao, X, Han, F, Feng, Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
7WWF
 
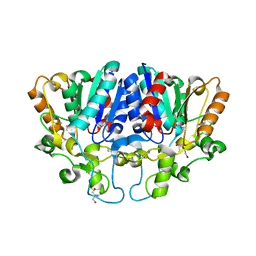 | | Crystal structure of BioH3 from Mycolicibacterium smegmatis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Esterase | | Authors: | Yang, J, Xu, Y.C, Gan, J.H, Feng, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-06 | | Last modified: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Three enigmatic BioH isoenzymes are programmed in the early stage of mycobacterial biotin synthesis, an attractive anti-TB drug target.
Plos Pathog., 18, 2022
|
|
4QGU
 
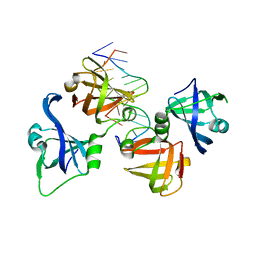 | | protein domain complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | Authors: | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | Deposit date: | 2014-05-25 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
8H97
 
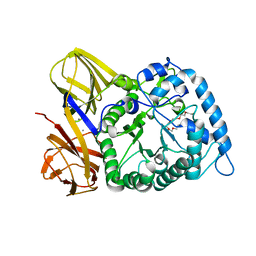 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
8HDJ
 
 | |
8WBO
 
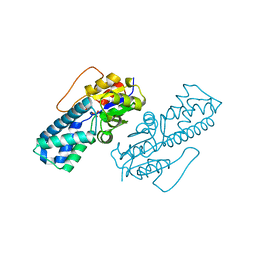 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D18N complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBS
 
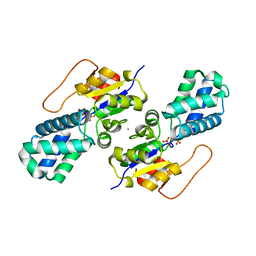 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L]-D48N complexed with sulfate ions | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBR
 
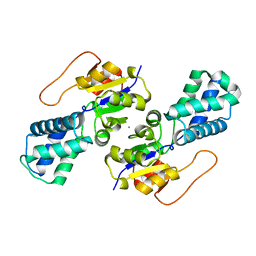 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBK
 
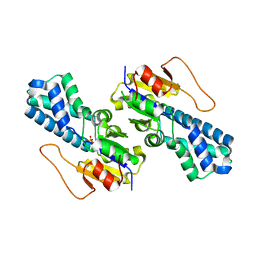 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBT
 
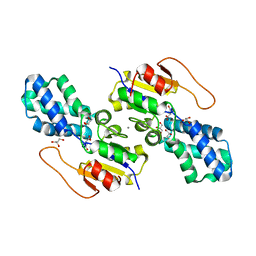 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] mutant D48N complexed with L-TA | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, GLYCEROL, ... | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBP
 
 | |
8WBQ
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant E212Q complexed with L-TA. | | Descriptor: | Epoxide hydrolase, L(+)-TARTARIC ACID | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBN
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193N | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBL
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
