4YU9
 
 | | Crystal Structure of double mutant Y115E Y117E human Glutaminyl Cyclase | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Di Pisa, F, Pozzi, C, Benvenuti, M, Mangani, S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The soluble Y115E-Y117E variant of human glutaminyl cyclase is a valid target for X-ray and NMR screening of inhibitors against Alzheimer disease.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3P43
 
 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4YWY
 
 | | Crystal Structure of double mutant Y115E Y117E human Glutaminyl Cyclase in complex with inhibitor PBD-150 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CHLORIDE ION, ... | | Authors: | Di Pisa, F, Pozzi, C, Benvenuti, M, Mangani, S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The soluble Y115E-Y117E variant of human glutaminyl cyclase is a valid target for X-ray and NMR screening of inhibitors against Alzheimer disease.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3OQ2
 
 | | Structure of a CRISPR associated protein Cas2 from Desulfovibrio vulgaris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Samai, P, Smith, P, Shuman, S. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
4XBA
 
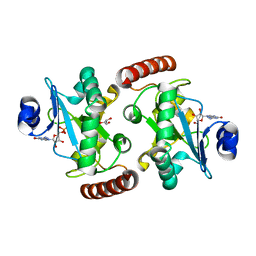 | | Hnt3 | | Descriptor: | Aprataxin-like protein, GLYCEROL, GUANOSINE, ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2014-12-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
4YKL
 
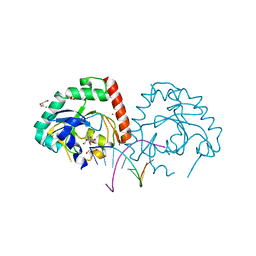 | | Hnt3 in complex with DNA and guanosine | | Descriptor: | Aprataxin-like protein, CHLORIDE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2HEG
 
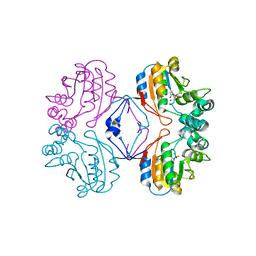 | | Phospho-Aspartyl Intermediate Analogue of Apha class B acid phosphatase/phosphotransferase | | Descriptor: | Class B acid phosphatase, MAGNESIUM ION | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-06-21 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
3P98
 
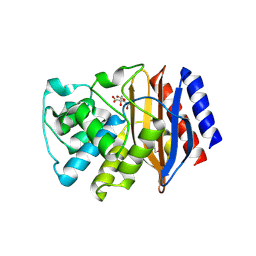 | | The crystal structure of the extended spectrum beta-lactamase TEM-72 reveals inhibition by citrate | | Descriptor: | Beta-lactamase TEM-72, CITRIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Rossolini, G.M, Mangani, S. | | Deposit date: | 2010-10-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the extended-spectrum [beta]-lactamase TEM-72 inhibited by citrate
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3P4H
 
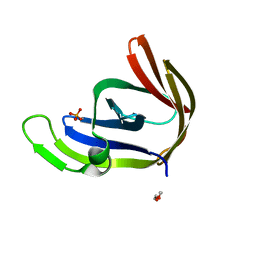 | | Structures of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Zhu, H, Shuman, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
2HF7
 
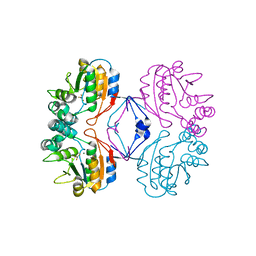 | | Transition State Analogue of AphA class B Acid Phosphatase/Phosphotransferase (Aluminium Fluoride Complex) | | Descriptor: | ALUMINUM FLUORIDE, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-06-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: |
|
|
1UN1
 
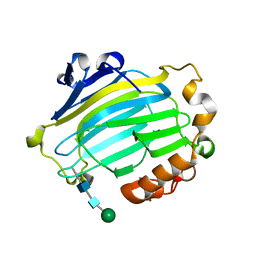 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1BZQ
 
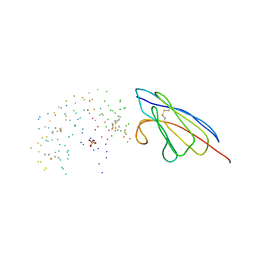 | | COMPLEX OF A DROMEDARY SINGLE-DOMAIN VHH ANTIBODY FRAGMENT WITH RNASE A | | Descriptor: | PHOSPHATE ION, PROTEIN (ANTIBODY CAB-RN05), PROTEIN (RNASE A) | | Authors: | Decanniere, K, Desmyter, A, Gahroudhi, M, Lauwereys, M, Muyldermans, S, Wyns, L. | | Deposit date: | 1998-11-03 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops.
Structure Fold.Des., 7, 1999
|
|
3KQ5
 
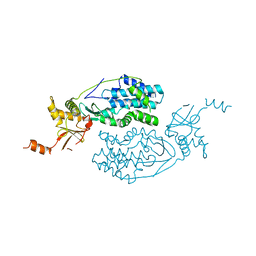 | | Crystal structure of an uncharacterized protein from Coxiella burnetii | | Descriptor: | Hypothetical cytosolic protein | | Authors: | Bonanno, J.B, Freeman, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-17 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Coxiella burnetii
To be Published
|
|
3PQV
 
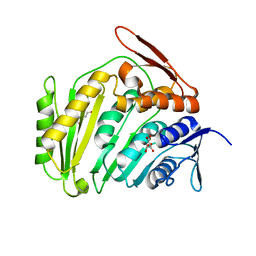 | | Cyclase homolog | | Descriptor: | D(-)-TARTARIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tanaka, N, Smith, P, Shuman, S. | | Deposit date: | 2010-11-27 | | Release date: | 2011-04-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis.
Rna, 17, 2011
|
|
3TTJ
 
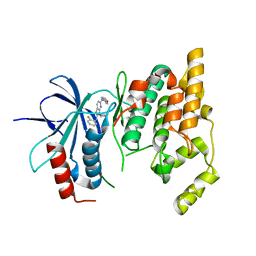 | | Crystal Structure of JNK3 complexed with CC-359, a JNK inhibitor for the prevention of ischemia-reperfusion injury | | Descriptor: | 9-cyclopentyl-N~8~-(2-fluorophenyl)-N~2~-(4-methoxyphenyl)-9H-purine-2,8-diamine, Mitogen-activated protein kinase 10 | | Authors: | Plantevin-Krenitsky, V, Delgado, M, Nadolny, L, Sahasrabudhe, K, Ayala, S, Clareen, S, Hilgraf, R, Albers, R, Kois, A, Hughes, K, Wright, J, Nowakowski, J, Sudbeck, E, Ghosh, S, Bahmanyar, S, Chamberlain, P, Muir, J, Cathers, B.E, Giegel, D, Xu, L, Celeridad, M, Moghaddam, M, Khatsenko, O, Omholt, P, Katz, J, Pai, S, Fan, R, Tang, Y, Shirley, M.A, Benish, B, Blease, K, Raymon, H, Bhagwat, S, Bennett, B, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopurine based JNK inhibitors for the prevention of ischemia reperfusion injury.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2LMK
 
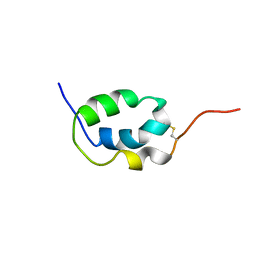 | | Solution Structure of Mouse Pheromone ESP1 | | Descriptor: | Exocrine gland-secreting peptide 1 | | Authors: | Yoshinaga, S, Sato, T, Hirakane, M, Esaki, K, Hamaguchi, T, Haga-Yamanaka, S, Tsunoda, M, Kimoto, H, Shimada, I, Touhara, K, Terasawa, H. | | Deposit date: | 2011-12-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mouse Sex Peptide Pheromone ESP1 Reveals a Molecular Basis for Specific Binding to the Class-C G-Protein-Coupled Vomeronasal Receptor
J.Biol.Chem., 2013
|
|
1S68
 
 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
1SXS
 
 | | Reduced bovine superoxide dismutase at pH 5.0 complexed with thiocyanate | | Descriptor: | CALCIUM ION, COPPER (II) ION, PROTEIN (CU-ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Mangani, S. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site
J.Biol.Inorg.Chem., 3, 1998
|
|
1SXB
 
 | | CRYSTAL STRUCTURE OF REDUCED BOVINE ERYTHROCYTE SUPEROXIDE DISMUTASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Rypniewski, W.R, Mangani, S, Bruni, B, Orioli, P, Casati, M, Wilson, K.S. | | Deposit date: | 1995-03-17 | | Release date: | 1995-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of reduced bovine erythrocyte superoxide dismutase at 1.9 A resolution.
J.Mol.Biol., 251, 1995
|
|
