4PPH
 
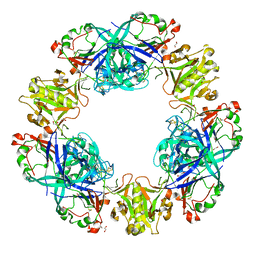 | | Crystal structure of conglutin gamma, a unique basic 7S globulin from lupine seeds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czubinski, J, Barciszewski, J, Gilski, M, Lampart-Szczapa, E, Jaskolski, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structure of gamma-conglutin: insight into the quaternary structure of 7S basic globulins from legumes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2RSM
 
 | | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65 (ICT2) | | Descriptor: | Probable peptide chain release factor C12orf65 homolog, mitochondrial | | Authors: | Enomoto, M, Tochio, N, Tomizawa, T, Koshiba, S, Guntert, P, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65.
Proteins, 2012
|
|
5ZSU
 
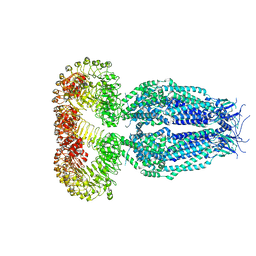 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1UDM
 
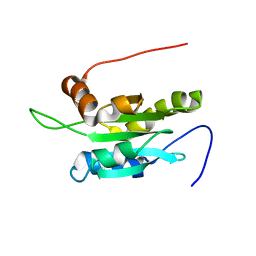 | | Solution structure of Coactosin-like protein (Cofilin family) from Mus Musculus | | Descriptor: | Coactosin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
7DC8
 
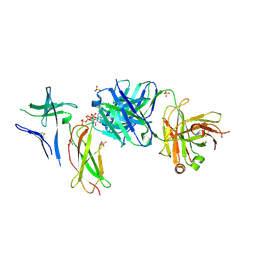 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7DC7
 
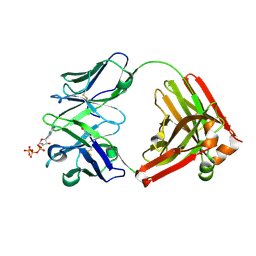 | | Crystal structure of D12 Fab-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | Authors: | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
1WGU
 
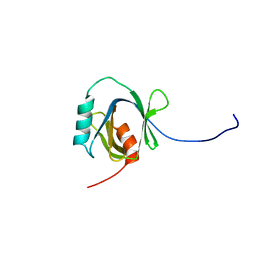 | | Solution Structure of the C-terminal Phosphotyrosine Interaction Domain of APBB2 from Mouse | | Descriptor: | amyloid beta (A4) precursor protein-binding, family B, member 2 | | Authors: | Li, H, Hayashi, F, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
7VTN
 
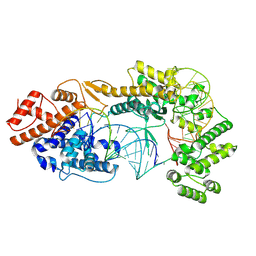 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
1X65
 
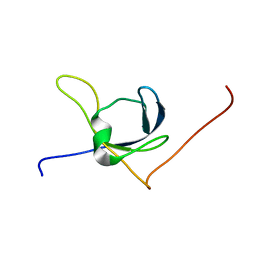 | | Solution structure of the third cold-shock domain of the human KIAA0885 protein (UNR PROTEIN) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YSZ
 
 | | Solution structure of the chimera of the C-terminal PID domain of Fe65L and the C-terminal tail peptide of APP | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2 and Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
5X7K
 
 | |
4GEL
 
 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEN
 
 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEM
 
 | | Crystal structure of Zucchini (K171A) | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
2YS5
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2YT0
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2YT1
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
6LN5
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
8JL8
 
 | | Crystal structure of the collagen binding domain of Cnm from Streptococcus mutans | | Descriptor: | Collagen-binding adhesin, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S.-i, Hirata, A, Takano, K. | | Deposit date: | 2023-06-02 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure, Stability and Binding Properties of Collagen-Binding Domains from Streptococcus mutans.
Chemistry, 5, 2023
|
|
6LN9
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class2) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN7
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class3) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN8
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class1) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
1WWY
 
 | | Solution structure of the DUF1000 domain of a thioredoxin-like protein 1 | | Descriptor: | Thioredoxin-like protein 1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal DUF1000 domain of the human thioredoxin-like 1 protein.
Proteins, 78, 2010
|
|
6LN6
 
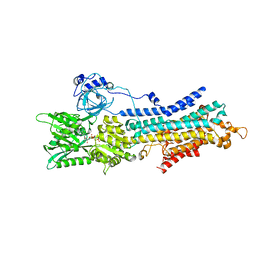 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class2) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LLE
 
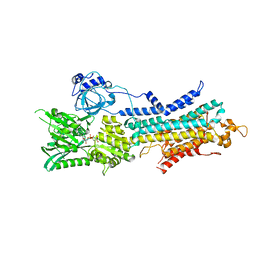 | | CryoEM structure of SERCA2b WT in E1-2Ca2+-AMPPCP state. | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
