4JGZ
 
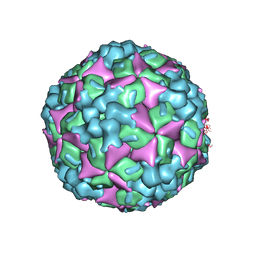 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4F5Y
 
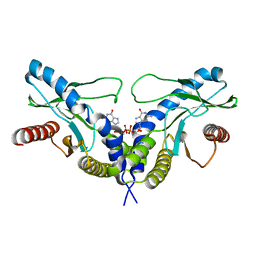 | | Crystal structure of human STING CTD complex with C-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
4JGY
 
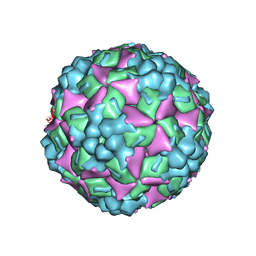 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4IU9
 
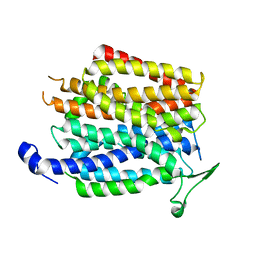 | | Crystal structure of a membrane transporter | | Descriptor: | Nitrite extrusion protein 2 | | Authors: | Yan, H, Huang, W, Yan, C, Gong, X, Jiang, S, Zhao, Y, Wang, J, Shi, Y. | | Deposit date: | 2013-01-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure and mechanism of a nitrate transporter.
Cell Rep, 3, 2013
|
|
3EB0
 
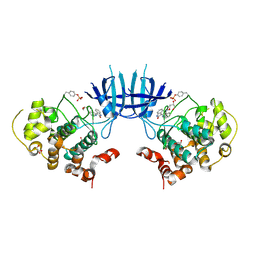 | | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, GLYCEROL, Putative uncharacterized protein | | Authors: | Wernimont, A.K, Fedorov, O, Lam, A, Ali, A, Zhao, Y, Lew, J, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804
TO BE PUBLISHED
|
|
1AAX
 
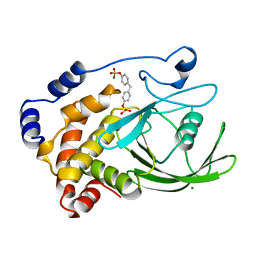 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO BIS(PARA-PHOSPHOPHENYL)METHANE (BPPM) MOLECULES | | Descriptor: | 4-PHOSPHONOOXY-PHENYL-METHYL-[4-PHOSPHONOOXY]BENZEN, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Puius, Y.A, Zhao, Y, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
3E95
 
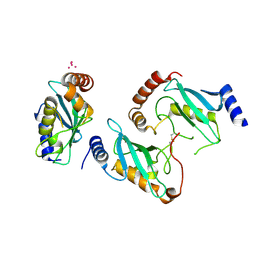 | | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carrier protein, Ubiquitin-conjugating enzyme E2 | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Brokx, S, Lin, Y.H, Zhao, Y, Lew, J, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, BOuntra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Qiu, W, Brand, V.B, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a
TO BE PUBLISHED
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
3FB3
 
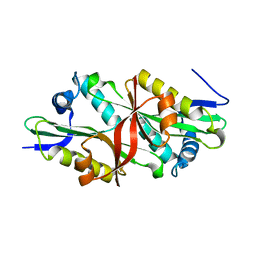 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
4I5X
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
3F9R
 
 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
3B6N
 
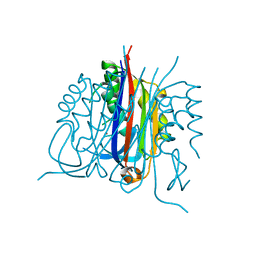 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax.
To be Published
|
|
4WKH
 
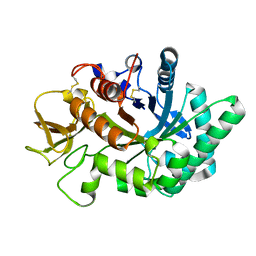 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3CKE
 
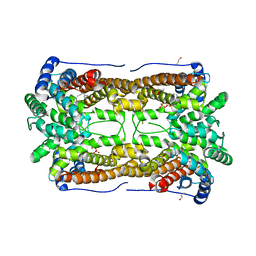 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
3BE4
 
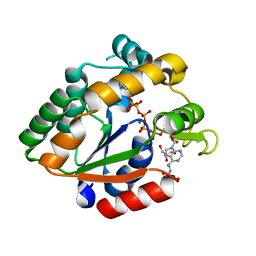 | | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, GLYCEROL, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360.
To be Published
|
|
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3I3G
 
 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
3BKP
 
 | | Crystal structure of the Toxoplasma gondii cyclophilin, 49.m03261 | | Descriptor: | Cyclophilin, GLYCEROL | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Toxoplasma gondii cyclophilin, 49.m03261.
To be Published
|
|
3BO7
 
 | | Crystal structure of Toxoplasma gondii peptidyl-prolyl cis-trans isomerase, 541.m00136 | | Descriptor: | 1,2-ETHANEDIOL, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE CYCLOPHILIN-TYPE, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-17 | | Release date: | 2008-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Peptidyl-Prolyl Cis-Trans Isomerase, 541.M00136.
To be Published
|
|
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3HJC
 
 | | Crystal structure of the carboxy-terminal domain of HSP90 from Leishmania major, LmjF33.0312 | | Descriptor: | Heat shock protein 83-1, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Walker, J, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the middle and carboxy-terminal domain of HSP90 from Leishmania major, LMJF33.0312
To be Published
|
|
4ZEL
 
 | | Human dopamine beta-hydroxylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Vendelboe, T.V, Harris, P, Christensen, H.E.M, Harlos, K, Walter, T, Zhao, Y, Omari, K. | | Deposit date: | 2015-04-20 | | Release date: | 2016-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human dopamine beta-hydroxylase at 2.9 angstrom resolution.
Sci Adv, 2, 2016
|
|
