4QTO
 
 | | 1.65 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-modified Cys289 and PEG molecule in active site | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P9H
 
 | |
4S04
 
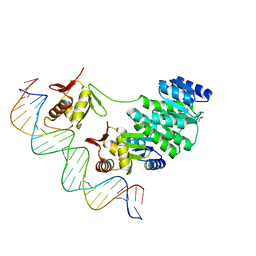 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (25-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
7KXJ
 
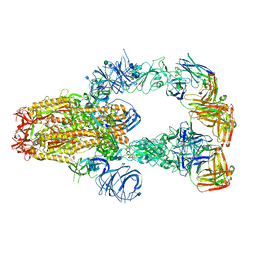 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
7KXK
 
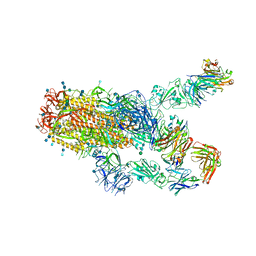 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 2-"up"-1-"down" conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7MQQ
 
 | |
8JYE
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYA
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYF
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with DMAPP | | Descriptor: | Butyrophylin 3, DIMETHYLALLYL DIPHOSPHATE, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYC
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with DMAPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JY9
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYB
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 | | Descriptor: | Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
5YU9
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with Ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation.
Oncotarget, 7, 2016
|
|
8SUX
 
 | | Structure of E. coli PtuA hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
8IZE
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IZG
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 5-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IXV
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A in complex with 2Cl-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IH4
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A2 Mutant | | Descriptor: | Butyrophilin subfamily 2 member A2 | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IGT
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A1 | | Descriptor: | Butyrophilin subfamily 2 member A1, ZINC ION | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-21 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
2O1M
 
 | | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572 | | Descriptor: | Probable amino-acid ABC transporter extracellular-binding protein ytmK, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572.
To be Published
|
|
2OBK
 
 | | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens. Northeast Structural Genomics Consortium target PlR6. | | Descriptor: | SelT/selW/selH selenoprotein domain | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens
To be Published
|
|
4B8T
 
 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
5J87
 
 | |
