9CID
 
 | |
9CIF
 
 | |
9CIC
 
 | |
9CIE
 
 | |
9CIG
 
 | |
7OP0
 
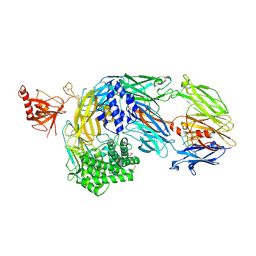 | | Crystal structure of complement C5 in complex with chemically synthesized K92 knob domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5 alpha chain, ... | | Authors: | Macpherson, A, van der Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Chemical Synthesis of Knob Domain Antibody Fragments.
Acs Chem.Biol., 16, 2021
|
|
2AXM
 
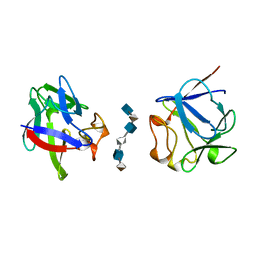 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Digabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-20 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
7JH6
 
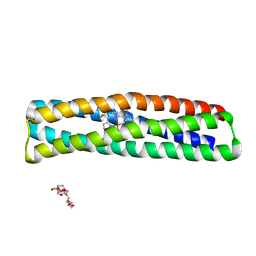 | | De novo designed two-domain di-Zn(II) and porphyrin-binding protein | | Descriptor: | NONAETHYLENE GLYCOL, Two-domain di-Zn(II) and porphyrin-binding protein, ZINC ION, ... | | Authors: | Schmidt, N, Liu, L, DeGrado, W.F. | | Deposit date: | 2020-07-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric cooperation in a de novo-designed two-domain protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QUG
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Gallium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING GA, ... | | Authors: | Moriwaki, Y, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-02-24 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus.
Biochemistry, 50, 2011
|
|
3QUH
 
 | |
5KZV
 
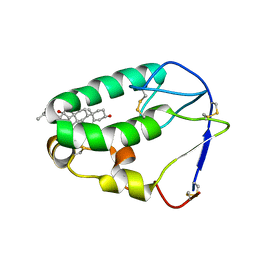 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in complex with 20(S)-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Smoothened | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
5KZY
 
 | |
5KZZ
 
 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in its apo-form | | Descriptor: | ACETATE ION, GLYCEROL, Smoothened, ... | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
3G2D
 
 | | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*G*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G00
 
 | | Mth0212 in complex with a 9bp blunt end dsDNA at 1.7 Angstrom | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*GP*TP*AP*TP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*UP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
7AD6
 
 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
7AD7
 
 | | Crystal structure of human complement C5 in complex with the K8 bovine knob domain peptide. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
4WYB
 
 | | Structure of the Bud6 flank domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Eck, M.J, Park, E, Zheng, W. | | Deposit date: | 2014-11-17 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Structure of a Bud6/Actin Complex Reveals a Novel WH2-like Actin Monomer Recruitment Motif.
Structure, 23, 2015
|
|
5B5K
 
 | | Crystal structure of Izumo1, the mammalian sperm ligand for egg Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Izumo sperm-egg fusion protein 1 | | Authors: | Nishimura, K, Han, L, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins.
Curr.Biol., 26, 2016
|
|
5XUQ
 
 | | Crystal structure of VDR-LBD complexed with an antagonist, 2-methylidene-19,26,27-trinor-22-(S)-butyl-1-hydroxy-25-oxo-25-(1H-pyrrol-2-yl)- vitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(1~{H}-pyrrol-2-yl)octan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-24 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent Vitamin D Receptor Antagonist
To Be Published
|
|
5GZ6
 
 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | Descriptor: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
 | | Structure of D-amino acid dehydrogenase in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
5GZ1
 
 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
7YXP
 
 | | Crystal structure of WT AncGR2-LBD WT bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
