2WZ9
 
 | | Crystal structure of the thioredoxin domain of human TXNL2 | | Descriptor: | GLUTAREDOXIN-3 | | Authors: | Vollmar, M, Johannson, C, Cocking, R, Pike, A.C.W, Muniz, J.R.C, Edwards, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Kavanagh, K.L. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Thioredoxin Domain of Human Txnl2
To be Published
|
|
4C45
 
 | | Crystal structure of human pterin-4-alpha-carbinolamine dehydratase 2 (PCBD2) | | Descriptor: | 1,2-ETHANEDIOL, PTERIN-4-ALPHA-CARBINOLAMINE DEHYDRATASE 2 | | Authors: | Kopec, J, Kiyani, W, Vollmar, M, Shrestha, L, Canning, P, von Delft, F, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W.W. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Human Pterin-4-Alpha- Carbinolamine Dehydratase 2 (Pcbd2)
To be Published
|
|
4CCZ
 
 | | Crystal structure of human 5-methyltetrahydrofolate-homocysteine methyltransferase, the homocysteine and folate binding domains | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, METHIONINE SYNTHASE | | Authors: | Vollmar, M, Kiyani, W, Krojer, T, Goubin, S, Burgess-Brown, N, von Delft, F, Oppermann, U, Edwards, A, Arrowsmith, C, Bountra, C, Yue, W.W. | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human 5-Methyltetrahydrofolate-Homocysteine Methyltransferase, the Homocysteine and Folate Binding Domains
To be Published
|
|
4BQM
 
 | | Crystal structure of human liver-type glutaminase, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTAMINASE LIVER ISOFORM, ... | | Authors: | Ferreira, I.M, Vollmar, M, Krojer, T, Strain-Damerell, C, Froese, S, Coutandin, D, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Dias, S.M.G, Ambrosio, A.L.B, Yue, W.W. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Human Liver-Type Glutaminase, Catalytic Domain
To be Published
|
|
2WMD
 
 | | Crystal structure of NmrA-like family domain containing protein 1 in complex with NADP and 2-(4-chloro-phenylamino)-nicotinic acid | | Descriptor: | 2-(4-CHLORO-PHENYLAMINO)-NICOTINIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NMRA-LIKE FAMILY DOMAIN CONTAINING PROTEIN 1 | | Authors: | Bhatia, C, Pilka, E, Niesen, F, Yue, W.W, Ugochukwu, E, Savitsky, P, Hozjan, V, Roos, A.K, Filippakopoulos, P, von Delft, F, Heightman, T, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Nmra-Like Family Domain Containing Protein 1
To be Published
|
|
2WNT
 
 | | Crystal Structure of the Human Ribosomal protein S6 kinase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, RIBOSOMAL PROTEIN S6 KINASE, ... | | Authors: | Muniz, J.R.C, Elkins, J.M, Wang, J, Ugochukwu, E, Salah, E, King, O, Picaud, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Ribosomal Protein S6 Kinase
To be Published
|
|
2X4D
 
 | | Crystal structure of human phospholysine phosphohistidine inorganic pyrophosphate phosphatase LHPP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vollmar, M, Gileadi, C, Guo, K, Savitsky, P, Muniz, J.R.C, Yue, W, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human Phospholysine Phosphohistidine Inorganic Pyrophosphate Phosphatase Lhpp
To be Published
|
|
2ATV
 
 | | The crystal structure of human RERG in the GDP bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-like estrogen-regulated growth inhibitor | | Authors: | Turnbull, A.P, Salah, E, Schoch, G, Elkins, J, Burgess, N, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human RERG in the GDP bound state
To be Published
|
|
2C60
 
 | | crystal structure of human mitogen-activated protein kinase kinase kinase 3 isoform 2 phox domain at 1.25 A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, HUMAN MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 3 ISOFORM 2 | | Authors: | Debreczeni, J.E, Salah, E, Papagrigoriou, E, Burgess, N, von Delft, F, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S. | | Deposit date: | 2005-11-04 | | Release date: | 2005-11-29 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Human Mitogen-Activated Protein Kinase Kinase Kinase 3 Isoform 2 Fox Domain at 1.25 A Resolution
To be Published
|
|
2C2H
 
 | | CRYSTAL STRUCTURE OF THE HUMAN RAC3 IN COMPLEX WITH GDP | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Yang, X, Zao, Y, Elkins, J, Gileadi, C, Burgess, N, Colebrook, S, Gileadi, O, Fedorov, O, Bunkoczi, G, von Delft, F, Doyle, D, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Human Rac3 in Complex with Gdp
To be Published
|
|
2C4K
 
 | | Crystal structure of human phosphoribosylpyrophosphate synthetase- associated protein 39 (PAP39) | | Descriptor: | PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE-ASSOCIATED PROTEIN 1, SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Kursula, P, Stenmark, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2005-10-20 | | Release date: | 2005-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Phosphoribosylpyrophosphate Synthetase-Associated Protein 39 (Pap39)
To be Published
|
|
2WWY
 
 | | Structure of human RECQ-like helicase in complex with a DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DA DG DC DG DT DC DG DA DG DA DT DC DCP)-3', ATP-DEPENDENT DNA HELICASE Q1, ... | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Chaikuad, A, Krojer, T, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recq1 Helicase-Driven DNA Unwinding, Annealing, and Branch Migration: Insights from DNA Complex Structures
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2C0C
 
 | | Structure of the MGC45594 gene product | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC BINDING ALCOHOL DEHYDROGENASE, DOMAIN CONTAINING 2 | | Authors: | Bunkoczi, G, Shafqat, N, Guo, K, Smee, C, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Gileadi, O, von Delft, F, Oppermann, U. | | Deposit date: | 2005-08-30 | | Release date: | 2005-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Structure of Mgc45594 Gene Product
To be Published
|
|
1ZSV
 
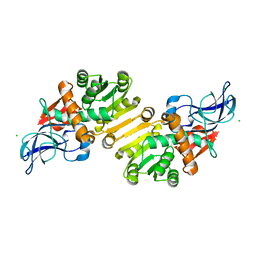 | | Crystal structure of human NADP-dependent leukotriene B4 12-hydroxydehydrogenase | | Descriptor: | CHLORIDE ION, NADP-dependent leukotriene B4 12-hydroxydehydrogenase | | Authors: | Turnbull, A.P, Johansson, C, Savitsky, P, Guo, K, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human NADP-dependent leukotriene B4 12-hydroxydehydrogenase
To be Published
|
|
1ZSX
 
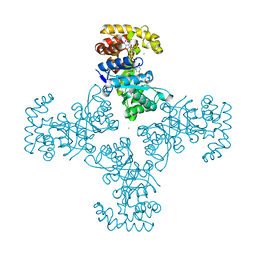 | | Crystal Structure Of Human Potassium Channel Kv Beta-subunit (KCNAB2) | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel beta-2 subunit | | Authors: | Wennerstrand, P, Johannsson, I, Ugochukwu, E, Kavanagh, K, Edwards, A, Arrowsmith, C, Williams, L, Sundstrom, M, Guo, K, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure Of Human Potassium Channel Kv Beta-subunit (KCNAB2)
To be Published
|
|
1SDI
 
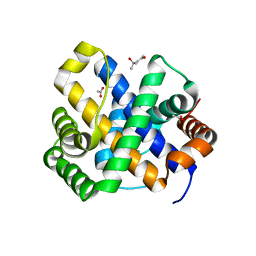 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
2CLS
 
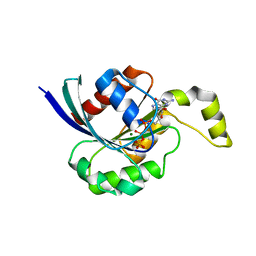 | | The crystal structure of the human RND1 GTPase in the active GTP bound state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHO6 | | Authors: | Pike, A.C.W, Yang, X, Colebrook, S, Gileadi, O, Sobott, F, Bray, J, Wen Hwa, L, Marsden, B, Zhao, Y, Schoch, G, Elkins, J, Debreczeni, J.E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State
To be Published
|
|
1ZSY
 
 | | The structure of human mitochondrial 2-enoyl thioester reductase (CGI-63) | | Descriptor: | GLYCEROL, MITOCHONDRIAL 2-ENOYL THIOESTER REDUCTASE, SULFATE ION | | Authors: | Lukacik, P, Shafqat, N, Kavanagh, K.L, Johansson, C, Smee, C, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of human mitochondrial 2-enoyl thioester reductase (CGI-63)
To be Published
|
|
4O38
 
 | | Crystal structure of the human cyclin G associated kinase (GAK) | | Descriptor: | Cyclin-G-associated kinase, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, R, Hatzos-Skintges, C, Weger, A, Chaikuad, A, Eswaran, J, Fedorov, O, King, O, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the human cyclin G associated kinase (GAK)
To be Published
|
|
1T57
 
 | | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved Protein MTH1675, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION | | Authors: | Kim, Y, Joachimiak, A, Saridakis, V, Xu, X, Arrowsmith, C.H, Christendat, D, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-03 | | Release date: | 2004-08-03 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum
To be Published
|
|
2C46
 
 | | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | | Descriptor: | MRNA CAPPING ENZYME | | Authors: | Debreczeni, J, Johansson, C, Longman, E, Gileadi, O, SavitskySmee, P, Smee, C, Bunkoczi, G, Ugochukwu, E, von Delft, F, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-15 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Human RNA Guanylyltransferase and 5'-Phosphatase
To be Published
|
|
2AW5
 
 | | Crystal structure of a human malic enzyme | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Papagrigoriou, E, Berridge, G, Smee, C, Bray, J, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Gileadi, O, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a human malic enzyme
To be published
|
|
2CFY
 
 | | Crystal structure of human thioredoxin reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE 1 | | Authors: | Debreczeni, J.E, Johansson, C, Kavanagh, K, Savitsky, P, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2006-02-26 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Thioredoxin Reductase 1
To be Published
|
|
1Z6Z
 
 | | Crystal Structure of Human Sepiapterin Reductase in complex with NADP+ | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Ugochukwu, E, Kavanagh, K, Ng, S, Arrowsmith, C, Edwards, A, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Sepiapterin Reductase
To be Published
|
|
2C9H
 
 | | Structure of mitochondrial beta-ketoacyl synthase | | Descriptor: | MITOCHONDRIAL BETA-KETOACYL SYNTHASE, NICKEL (II) ION | | Authors: | Bunkoczi, G, Wu, X, Smee, C, Gileadi, O, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Oppermann, U. | | Deposit date: | 2005-12-12 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Mitochondrial Beta-Ketoacyl Synthase
To be Published
|
|
