2JR7
 
 | | Solution structure of human DESR1 | | Descriptor: | DPH3 homolog, ZINC ION | | Authors: | Wu, F, Wu, J, Shi, Y. | | Deposit date: | 2007-06-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human DESR1, a CSL zinc-binding protein
Proteins, 71, 2008
|
|
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | Authors: | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
7E56
 
 | |
8JF4
 
 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | Authors: | Zhu, C.J. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89288354 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JG8
 
 | |
7DM2
 
 | | crystal structure of the M. tuberculosis phosphate ABC transport receptor PstS-1 in complex with Fab p4-170 | | Descriptor: | PHOSPHATE ION, Phosphate-binding protein PstS 1, heavy chain, ... | | Authors: | Ma, B, Freund, N, Xiang, Y. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Nat Commun, 12, 2021
|
|
7DM1
 
 | | crystal structure of the M.tuberculosis phosphate ABC transport receptor PstS-1 in complex with Fab p4-36 | | Descriptor: | PHOSPHATE ION, Phosphate-binding protein PstS 1, heavy chain, ... | | Authors: | Ma, B, Freund, N, Xiang, Y. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Nat Commun, 12, 2021
|
|
7W5M
 
 | |
7KLZ
 
 | |
6LE0
 
 | |
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
7VCQ
 
 | |
7VCL
 
 | |
6MIE
 
 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6ZYT
 
 | | Monomeric streptavidin with a conjugated biotinylated pyrrolidine | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, SULFATE ION, Streptavidin/Rhizavidin Hybrid | | Authors: | Nodling, A.R, Lipka-Lloyd, M, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V6D
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) cross-linked with AZT-terminated DNA | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7048 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7CN9
 
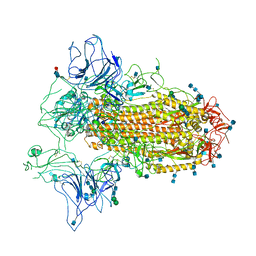 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
3V4I
 
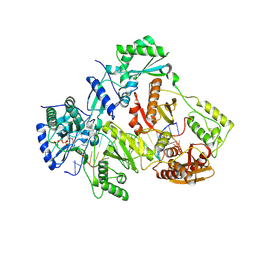 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and AZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7983 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3A5U
 
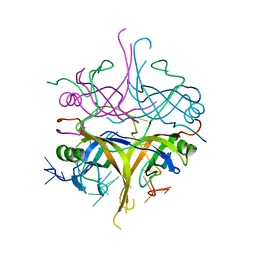 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
2HX4
 
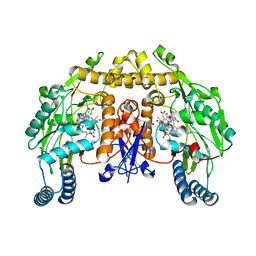 | | Rat nNOS heme domain complexed with 4-N-(Nw-nitro-L-argininyl)-trans-4-hydroxyamino-L-proline amide | | Descriptor: | (4R)-4-(HYDROXY{N~5~-[IMINO(NITROAMINO)METHYL]-L-ORNITHYL}AMINO)-L-PROLINAMIDE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX2
 
 | | Bovine eNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | Descriptor: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX3
 
 | | Rat nNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | Descriptor: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2006-08-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
7CNA
 
 | | Crystal structure of Spindlin1/C11orf84 complex bound to histone H3K4me3K9me3 peptide | | Descriptor: | ALA-ARG-THR-M3L-GLN-THR-ALA-ARG-M3L-SER-GLY, ALA-ARG-THR-M3L-GLN-THR-ALA-ARG-M3L-SER-THR, BENZAMIDINE, ... | | Authors: | Qian, C.M. | | Deposit date: | 2020-07-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural mechanism of bivalent histone H3K4me3K9me3 recognition by the Spindlin1/C11orf84 complex in rRNA transcription activation.
Nat Commun, 12, 2021
|
|
7WK1
 
 | | Mouse Pendrin bound chloride in inward state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
