7WGA
 
 | |
1SDA
 
 | | CRYSTAL STRUCTURE OF PEROXYNITRITE-MODIFIED BOVINE CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Smith, C.D, Carson, M, Van Der Woerd, M, Chen, J, Ischiropoulos, H, Beckman, J.S. | | Deposit date: | 1993-01-13 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of peroxynitrite-modified bovine Cu,Zn superoxide dismutase.
Arch.Biochem.Biophys., 299, 1992
|
|
3TUB
 
 | |
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6BE3
 
 | |
6BE4
 
 | |
3FZO
 
 | | Crystal Structure of PYK2-Apo, Proline-rich Tyrosine Kinase | | Descriptor: | Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZR
 
 | | Crystal structure of PYK2 complexed with PF-431396 | | Descriptor: | N-methyl-N-{2-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]phenyl}methanesulfonamide, PHOSPHATE ION, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZS
 
 | | Crystal Structure of PYK2 complexed with BIRB796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
4DF6
 
 | |
6HLK
 
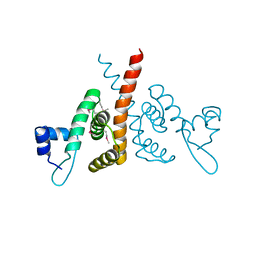 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC) | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-11 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
6DI7
 
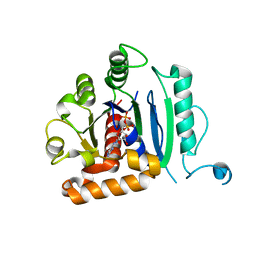 | | Vps1 GTPase-BSE fusion complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative sorting protein | | Authors: | Varlakhanova, N.V, Brady, T.M, Ford, M.G.J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
3FZP
 
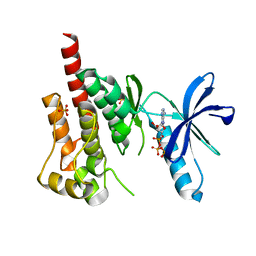 | | Crystal structure of PYK2 complexed with ATPgS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZT
 
 | | Crystal structure of PYK2 complexed with PF-4618433 | | Descriptor: | 1-[5-tert-butyl-2-(4-methylphenyl)-1,2-dihydro-3H-pyrazol-3-ylidene]-3-{3-[(pyridin-3-yloxy)methyl]-1H-pyrazol-5-yl}urea, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
4HCX
 
 | |
1VKA
 
 | | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Dailey, T.A, Mayer, M.R, Rose, J.P, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment
TO BE PUBLISHED
|
|
6E5G
 
 | |
3EBX
 
 | | REFINEMENT AT 1.4 ANGSTROMS RESOLUTION OF A MODEL OF ERABUTOXIN B. TREATMENT OF ORDERED SOLVENT AND DISCRETE DISORDER | | Descriptor: | ERABUTOXIN B, SULFATE ION | | Authors: | Smith, J.L, Corfield, P.W.R, Hendrickson, W.A, Low, B.W. | | Deposit date: | 1988-01-15 | | Release date: | 1988-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Refinement at 1.4 A resolution of a model of erabutoxin b: treatment of ordered solvent and discrete disorder.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
5TBD
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb4D11) in complex with 3D7-MSP2 215-222 | | Descriptor: | Fv fragment (mAb4D1) heavy chain, Merozoite surface protein 2 | | Authors: | Seow, J, Morales, R.A.V, MacRaild, C.A, Bankala, K, Drinkwater, N, Dingjan, T, Jaipuria, G, Wilde, K, Anders, R.F, Atreya, H.S, Christ, D, McGowan, S, Norton, R.S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Characterisation of a Key Epitope in the Conserved C-Terminal Domain of the Malaria Vaccine Candidate MSP2.
J. Mol. Biol., 429, 2017
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
3M6H
 
 | | Crystal Structure of Post-isomerized Ertapenem Covalent Adduct with TB B-lactamase | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
4MC3
 
 | | Hedycaryol synthase in complex with Nerolidol | | Descriptor: | (3R,6E)-3,7,11-trimethyldodeca-1,6,10-trien-3-ol, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
4JLF
 
 | | Inhibitor resistant (R220A) substitution in the Mycobacterium tuberculosis beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Kurz, S, Blanchard, J, Bonomo, R. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis beta-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of beta-lactam-beta-lactamase inhibitor combinations.
Antimicrob.Agents Chemother., 57, 2013
|
|
2GV9
 
 | |
1BW9
 
 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND PHENYLPYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYLPYRUVIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
