3KHZ
 
 | |
1W1T
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(His-L-Pro) at 1.9 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
2QRK
 
 | | Crystal Structure of AMP-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Saccharopine dehydrogenase [NAD+, L-lysine-forming | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
3KLS
 
 | | Structure of complement C5 in complex with SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KHX
 
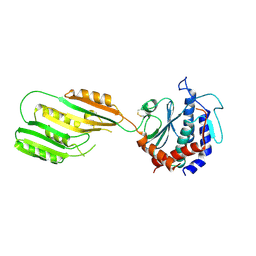 | |
2QWX
 
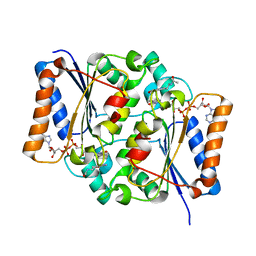 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QX4
 
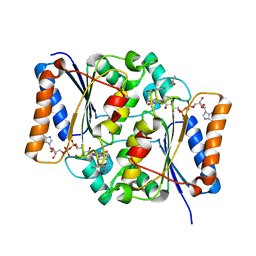 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
3KM9
 
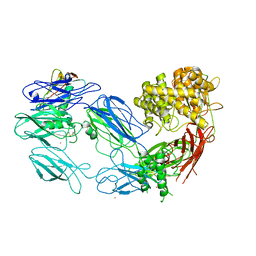 | | Structure of complement C5 in complex with the C-terminal beta-grasp domain of SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KPL
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPS
 
 | | Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
2QEF
 
 | | X-ray structure of 7-deaza-dG and Z3dU modified duplex CGCGAATXCZCG | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*(ZDU)P*DCP*(7GU)P*DCP*DG)-3') | | Authors: | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2007-06-25 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
2QGZ
 
 | | Crystal structure of a putative primosome component from Streptococcus pyogenes serotype M3. Northeast Structural Genomics target DR58 | | Descriptor: | PHOSPHATE ION, Putative primosome component | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative primosome component from Streptococcus pyogenes serotype M3.
To be Published
|
|
2QPD
 
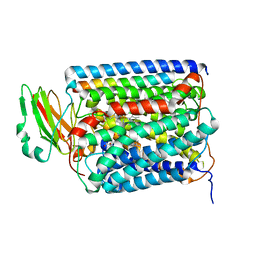 | | An unexpected outcome of surface-engineering an integral membrane protein: Improved crystallization of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Luna, V.M, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba(3) from Thermus thermophilus.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2QRJ
 
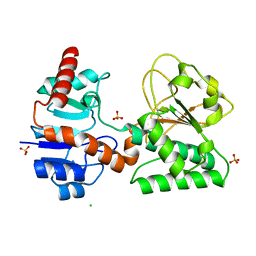 | | Crystal Structure of Sulfate-bound Saccharopine Dehydrogenase (L-Lys Forming) from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, SULFATE ION, Saccharopine dehydrogenase, ... | | Authors: | Andi, B, Xu, H, Cook, P.F, West, A.H. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Ligand-Bound Saccharopine Dehydrogenase from Saccharomyces cerevisiae
Biochemistry, 46, 2007
|
|
1WA8
 
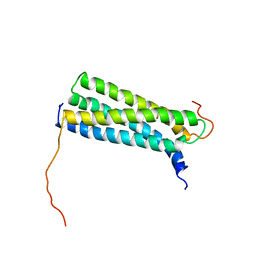 | | Solution Structure of the CFP-10.ESAT-6 Complex. Major Virulence Determinants of Pathogenic Mycobacteria | | Descriptor: | 6 KDA EARLY SECRETORY ANTIGENIC TARGET (ESAT-6), ESAT-6 LIKE PROTEIN ESXB | | Authors: | Renshaw, P.S, Lightbody, K.L, Veverka, V, Muskett, F.W, Kelly, G, Frenkiel, T.A, Gordon, S.V, Hewinson, R.G, Burke, B, Norman, J, Williamson, R.A, Carr, M.D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-25 | | Release date: | 2005-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of the Complex Formed by the Tuberculosis Virulence Factors Cfp-10 and Esat-6
Embo J., 24, 2005
|
|
3KZ4
 
 | | Crystal Structure of the Rotavirus Double Layered Particle | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Mcclain, B, Settembre, E.C, Bellamy, A.R, Harrison, S.C. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution.
J.Mol.Biol., 397, 2010
|
|
3L3Q
 
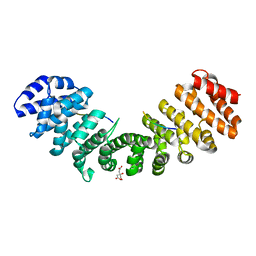 | | Mouse importin alpha-pepTM NLS peptide complex | | Descriptor: | CITRATE ANION, Importin subunit alpha-2, pepTM | | Authors: | Takeda, A.A.S, Kobe, B, Fontes, M.R.M. | | Deposit date: | 2009-12-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening.
J.Biol.Chem., 285, 2010
|
|
3KTB
 
 | | Crystal Structure of Arsenical Resistance Operon Trans-acting Repressor from Bacteroides vulgatus ATCC 8482 | | Descriptor: | ACETIC ACID, Arsenical resistance operon trans-acting repressor, CALCIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-24 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Arsenical Resistance Operon Trans-acting Repressor from Bacteroides vulgatus ATCC 8482
To be Published
|
|
1WG0
 
 | | Structural comparison of Nas6p protein structures in two different crystal forms | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Nakamura, Y, Umehara, T, Tanaka, A, Horikoshi, M, Yokoyama, S, Padmanabhan, B, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural comparison of Nas6p protein structures in two different crystal forms
To be Published
|
|
2QSI
 
 | | Crystal structure of putative hydrogenase expression/formation protein hupG from Rhodopseudomonas palustris CGA009 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Putative hydrogenase expression/formation protein hupG | | Authors: | Nocek, B, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative hydrogenase expression/formation protein hupG from Rhodopseudomonas palustris CGA009.
To be Published
|
|
2QGG
 
 | | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248. Northeast Structural Genomics Consortium target AsR73. | | Descriptor: | 16S rRNA-processing protein rimM, POTASSIUM ION, UNKNOWN LIGAND | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248.
To be Published
|
|
2QGV
 
 | | Crystal structure of hydrogenase-1 operon protein hyaE from Shigella flexneri. Northeast Structural Genomics Consortium Target SfR170 | | Descriptor: | Hydrogenase-1 operon protein hyaE | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Benach, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hydrogenase-1 operon protein hyaE from Shigella flexneri.
To be Published
|
|
1WW6
 
 | | Agrocybe cylindracea galectin complexed with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
2QL7
 
 | | Crystal Structure of Caspase-7 with inhibitor AC-IEPD-CHO | | Descriptor: | CITRIC ACID, Caspase-7, Inhibitor AC-IEPD_CHO, ... | | Authors: | Agniswamy, J, Fang, B, Weber, I. | | Deposit date: | 2007-07-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
3KXL
 
 | | crystal structure of SsGBP mutation variant G235S | | Descriptor: | GTP-binding protein (HflX), THIOCYANATE ION | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
