9IWY
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with LK01003 | | Descriptor: | 2-cyclopentyl-~{N}-(6-propan-2-ylsulfonylquinolin-4-yl)-1,3-benzothiazol-5-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IX2
 
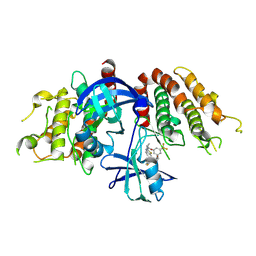 | | Crystal structure of the mouse RIP3 kinase domain in complexed with TAK-632 | | Descriptor: | N-{7-cyano-6-[4-fluoro-3-({[3-(trifluoromethyl)phenyl]acetyl}amino)phenoxy]-1,3-benzothiazol-2-yl}cyclopropanecarboxamide, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IWX
 
 | | Crystal structure of the mouse RIP3 kinase domain(R69H) in complexed with GSK'872 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3, ~{N}-(6-propan-2-ylsulfonylquinolin-4-yl)-1,3-benzothiazol-5-amine | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IWW
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with GSK'872 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3, ~{N}-(6-propan-2-ylsulfonylquinolin-4-yl)-1,3-benzothiazol-5-amine | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IWZ
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with GSK'843 | | Descriptor: | 3-(1,3-benzothiazol-5-yl)-7-(1,3-dimethyl-1H-pyrazol-5-yl)thieno[3,2-c]pyridin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IX1
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with PP2 | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IX0
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with GW'39B | | Descriptor: | 4-methyl-3-[(7-pyridin-2-ylquinolin-4-yl)amino]phenol, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
9IX3
 
 | | Crystal structure of the mouse RIP3 kinase domain in complexed with compound 18 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3, ~{N}-[4-[2-(cyclopropylcarbonylamino)pyridin-4-yl]oxy-2,3-dimethyl-phenyl]-1-(4-fluorophenyl)-2-oxidanylidene-pyridine-3-carboxamide | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-07-26 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
5B73
 
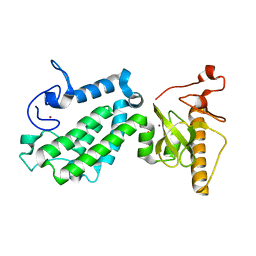 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GXR
 
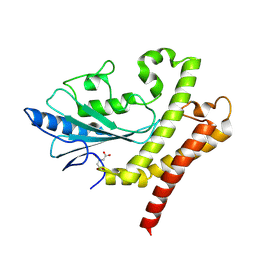 | | crystal structure of UBC domain of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O, CITRIC ACID | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the biochemical mechanism of the E2/E3 hybrid enzyme UBE2O.
Structure, 33, 2025
|
|
5GRS
 
 | | Complex structure of the fission yeast SREBP-SCAP binding domains | | Descriptor: | Sterol regulatory element-binding protein 1, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Qian, H.W, Wu, J.P, Yan, N. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization
Cell Res., 26, 2016
|
|
5GPD
 
 | |
8USZ
 
 | | Cryo-EM Structure of Full-Length Spike Protein of Omicron XBB.1.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Huynh, K.W, Chang, J.S, Fennell, K.F, Che, Y, Wu, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-12-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Preclinical characterization of the Omicron XBB.1.5-adapted BNT162b2 COVID-19 vaccine.
Npj Vaccines, 9, 2024
|
|
8GY6
 
 | |
3T9N
 
 | | Crystal structure of a membrane protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | Authors: | Yang, M, Zhang, X, Ge, J, Wang, J. | | Deposit date: | 2011-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
3TFQ
 
 | | Crystal structure of 11b-hsd1 double mutant (l262r, f278e) complexed with 8-{[(2-CYANOPYRIDIN-3-YL)METHYL]SULFANYL}-6-HYDROXY-3,4-DIHYDRO-1H-PYRANO[3,4-C]PYRIDINE-5-CARBONITRILE | | Descriptor: | 8-{[(2-cyanopyridin-3-yl)methyl]sulfanyl}-6-hydroxy-3,4-dihydro-1H-pyrano[3,4-c]pyridine-5-carbonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 3-hydroxy-4-cyano-isoquinolines as novel, potent, and selective inhibitors of human 11beta-hydroxydehydrogenase 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5MX7
 
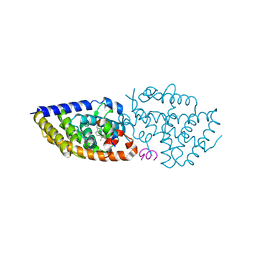 | | 1a,20S-dihydroxyvitamin D3 VDR complex | | Descriptor: | 1a,20S-dihydroxyvitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2017-01-21 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1 alpha,20S-Dihydroxyvitamin D3 Interacts with Vitamin D Receptor: Crystal Structure and Route of Chemical Synthesis.
Sci Rep, 7, 2017
|
|
5DJN
 
 | |
5YAX
 
 | | Crystal structure of a human neutralizing antibody bound to a HBV preS1 peptide | | Descriptor: | Large envelope protein, SODIUM ION, scFv1 antibody | | Authors: | Liu, X, Zheng, S, Ye, K, Sui, J. | | Deposit date: | 2017-09-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Elife, 6, 2017
|
|
7YW1
 
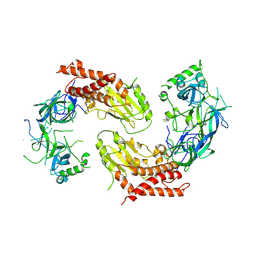 | | crystal structure of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structural insights into the biochemical mechanism of the E2/E3 hybrid enzyme UBE2O.
Structure, 33, 2025
|
|
4N4I
 
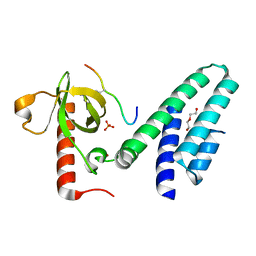 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4H
 
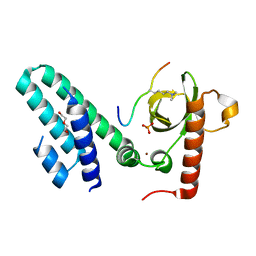 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
5TOZ
 
 | | JAK3 with covalent inhibitor PF-06651600 | | Descriptor: | 1-{(2S,5R)-2-methyl-5-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-10-19 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a JAK3-Selective Inhibitor: Functional Differentiation of JAK3-Selective Inhibition over pan-JAK or JAK1-Selective Inhibition.
ACS Chem. Biol., 11, 2016
|
|
