7Y3B
 
 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
 | | Crystal structure of TRIM7 bound to RACO-1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6KKR
 
 | | human KCC1 structure determined in KCl and detergent GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKT
 
 | | human KCC1 structure determined in KCl and lipid nanodisc | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKU
 
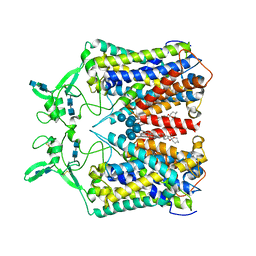 | | human KCC1 structure determined in NaCl and GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
3BHJ
 
 | | Crystal structure of human Carbonyl Reductase 1 in complex with glutathione | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, Carbonyl reductase [NADPH] 1, ... | | Authors: | Rauh, D, Bateman, R.L, Shokat, K.M. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Human carbonyl reductase 1 is an s-nitrosoglutathione reductase
J.Biol.Chem., 283, 2008
|
|
3BHM
 
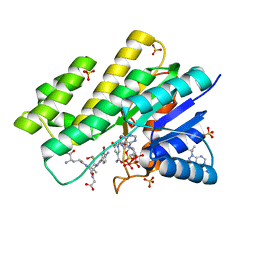 | | Crystal structure of human Carbonyl Reductase 1 in complex with S-hydroxymethylglutathione | | Descriptor: | 2-AMINO-4-[1-CARBOXYMETHYL-CARBAMOYL)-2-HYDROXYMETHYLSULFANYL-ETHYLCARBAMOYL]-BUTYRIC ACID, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, Carbonyl reductase [NADPH] 1, ... | | Authors: | Bateman, R.L, Rauh, D, Shokat, K.M. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human carbonyl reductase 1 is an s-nitrosoglutathione reductase
J.Biol.Chem., 283, 2008
|
|
3BRP
 
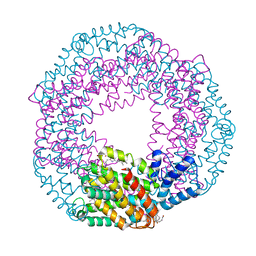 | | Crystal Structure of C-Phycocyanin from Galdieria sulphuraria | | Descriptor: | BILIVERDINE IX ALPHA, C-phycocyanin alpha chain, C-phycocyanin beta chain | | Authors: | Fromme, R. | | Deposit date: | 2007-12-21 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Galdieria sulphuraria at 1.85 A
To be Published
|
|
6E5P
 
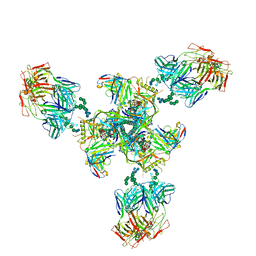 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | Descriptor: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-07-21 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
6EOJ
 
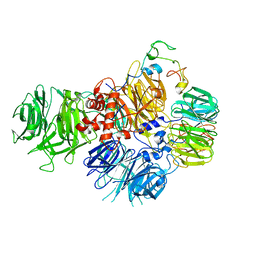 | | PolyA polymerase module of the cleavage and polyadenylation factor (CPF) from Saccharomyces cerevisiae | | Descriptor: | Polyadenylation factor subunit 2,Polyadenylation factor subunit 2, Protein CFT1, ZINC ION, ... | | Authors: | Casanal, A, Kumar, A, Hill, C.H, Emsley, P, Passmore, L.A. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Architecture of eukaryotic mRNA 3'-end processing machinery.
Science, 358, 2017
|
|
8J4A
 
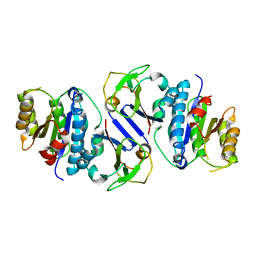 | | Crystal structure of OY phytoplasma SAP05 in complex with AtRPN10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J49
 
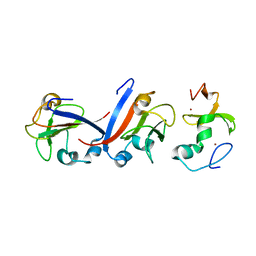 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL5 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J48
 
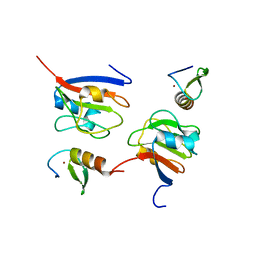 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J4B
 
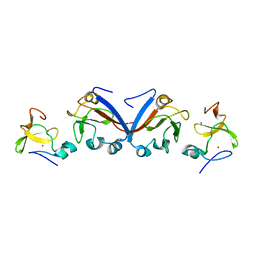 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL13 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 13A, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
6LAF
 
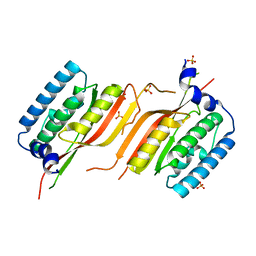 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | Descriptor: | Amuc_1100, SULFATE ION | | Authors: | Wang, J, Xiang, R, Zhang, M, Wang, M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
6EDD
 
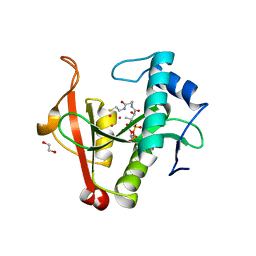 | | Crystal structure of a GNAT Superfamily PA3944 acetyltransferase in complex with CoA (P1 space group) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
6EDV
 
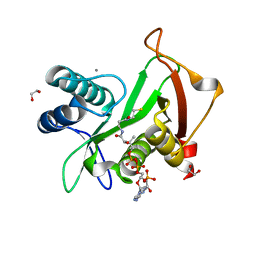 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | Authors: | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-12 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
6BR8
 
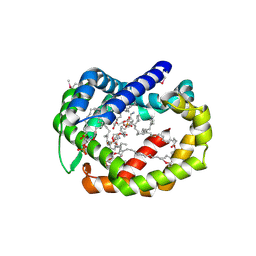 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BR9
 
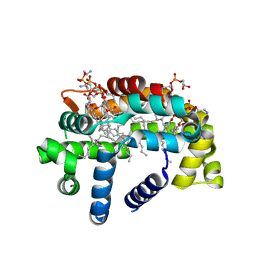 | | Structure of A6 reveals a novel lipid transporter | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, J, Peng, S, Pathak, P. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6LIR
 
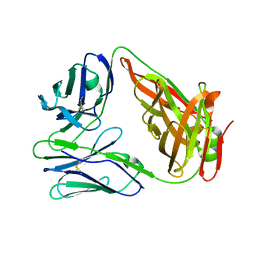 | | crystal structure of chicken TCR for 2.0 | | Descriptor: | TCR alpha chain, TCR beta chain | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural and Biophysical Insights into the TCR alpha beta Complex in Chickens.
Iscience, 23, 2020
|
|
6M67
 
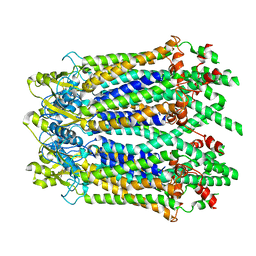 | | The Cryo-EM Structure of Human Pannexin 1 with D376E/D379E Mutation | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
7RFB
 
 | | Crystal structure of broadly neutralizing antibody mAb1198 in complex with Hepatitis C virus envelope glycoprotein E2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization.
Immunity, 55, 2022
|
|
7RFC
 
 | |
6M68
 
 | | The Cryo-EM Structure of Human Pannexin 1 in the Presence of CBX | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6M66
 
 | | The Cryo-EM Structure of Human Pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
