7B3O
 
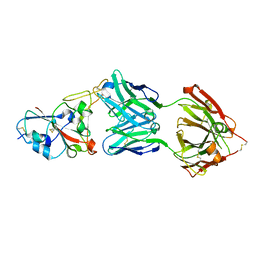 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
8QOY
 
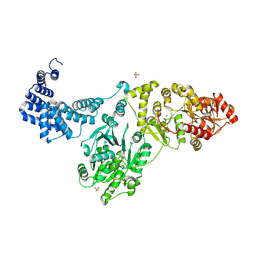 | | Capsular polysaccharide synthesis multienzyme of Actinobacillus Pleuropneumoniae serotype 3 | | Descriptor: | SULFATE ION, TagF-like capsule polymerase Cps3D, ZINC ION | | Authors: | Di Domenico, V, Litschko, C, Schulze, J, Bethe, A, Cifuente, J.O, Marina, A, Budde, I, Mast, T.A, Sulewska, M, Berger, M, Buettner, F, Gerardy-Schahn, R, Fiebig, T, Guerin, M.E. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transition transferases prime bacterial capsule polymerization.
Nat.Chem.Biol., 2024
|
|
5ML4
 
 | | The crystal structure of PDE6D in complex to inhibitor-7 | | Descriptor: | 4-[[[4-[(4-chlorophenyl)methyl-cyclopentyl-sulfamoyl]phenyl]sulfonyl-(piperidin-4-ylmethyl)amino]methyl]-2-(methylamino)benzoic acid, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Fansa, E.K, Martin-Gago, P, waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML6
 
 | | The crystal structure of PDE6D in complex to inhibitor-8 | | Descriptor: | 2-azanyl-4-[[[4-[(4-chlorophenyl)methyl-cyclopentyl-sulfamoyl]phenyl]sulfonyl-(piperidin-4-ylmethyl)amino]methyl]benzoic acid, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Fansa, E.K, Martin-gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML2
 
 | | The crystal structure of PDE6D in complex with inhibitor-3 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}1-[(4-chlorophenyl)methyl]-~{N}1-cyclopentyl-~{N}4-(phenylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML3
 
 | | The crystal structure of PDE6D in complex to Deltasonamide1 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}1-[(4-chlorophenyl)methyl]-~{N}1-cyclopentyl-~{N}4-[[2-(methylamino)pyrimidin-4-yl]methyl]-~{N}4-(piperidin-4-ylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ML8
 
 | | The crystal structure of PDE6D in complex to inhibitor-4 | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}4-cyclopentyl-~{N}1-(phenylmethyl)-~{N}1-(piperidin-4-ylmethyl)benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A PDE6 delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4TPZ
 
 | |
7AOS
 
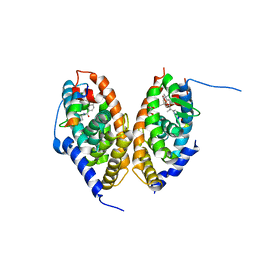 | | crystal structure of the RARalpha/RXRalpha ligand binding domain heterodimer in complex with a fragment of SRC1 coactivator | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, GLYCEROL, ... | | Authors: | le Maire, A, Guee, L, Bourguet, W. | | Deposit date: | 2020-10-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
4TO7
 
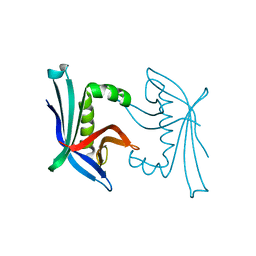 | |
7APO
 
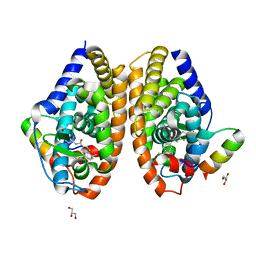 | | Crystal structure of RARalpha ligand binding domain in complex with a fragment of the TIF2 coactivator | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, GLYCEROL, Nuclear receptor coactivator 2, ... | | Authors: | le Maire, A, Guee, L, Bourguet, W. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
4TQ7
 
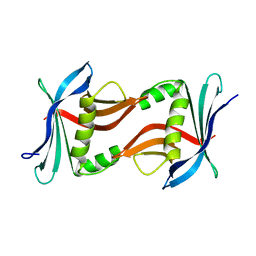 | |
4TTZ
 
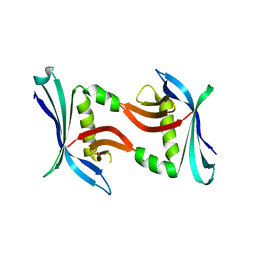 | |
4TTW
 
 | |
4TTX
 
 | |
4U2J
 
 | |
3ZGO
 
 | |
6P6G
 
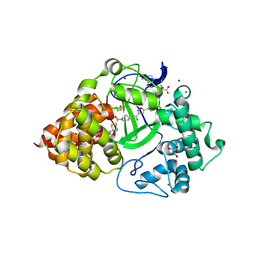 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P6K
 
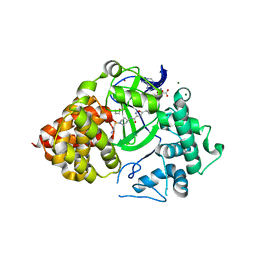 | |
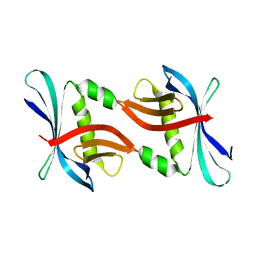
4TTY
 
 | |
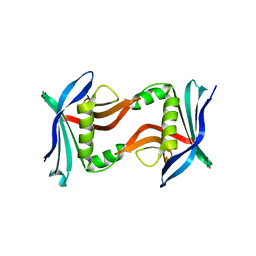
4U2I
 
 | |
7QUL
 
 | | Alcohol Dehydrogenase from Thauera aromatica K319A/K320A mutant | | Descriptor: | 1,2-ETHANEDIOL, 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, ZINC ION | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7QUY
 
 | | Alcohol Dehydrogenase from Thauera aromatica complexed with NADH | | Descriptor: | 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7A0Q
 
 | |
