4WXE
 
 | | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS
To be published
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
4X04
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CITROBACTER KOSERI (CKO_04899, TARGET EFI-510094) WITH BOUND D-glucuronate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, beta-D-glucopyranuronic acid, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CITROBACTER KOSERI (CKO_04899, TARGET EFI-510094) WITH BOUND D-glucuronate
To be published
|
|
4X9T
 
 | | Crystal structure of a TctC solute binding protein from Polaromonas (Bpro_3516, Target EFI-510338), no ligand | | Descriptor: | CHLORIDE ION, Uncharacterized protein UPF0065 | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of a TctC solute binding protein from Polaromonas (Bpro_3516, Target EFI-510338), no ligand
To be published
|
|
4XEQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (Deval_0042, TARGET EFI-510114) BOUND TO COPURIFIED (R)-PANTOIC ACID | | Descriptor: | PANTOATE, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-24 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (Deval_0042, TARGET EFI-510114) BOUND TO COPURIFIED (R)-PANTOIC ACID
To be published
|
|
4XHD
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN WITH COMPOUND-1 | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-cyclopropylacetamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR.
J.Mol.Biol., 427, 2015
|
|
4XF5
 
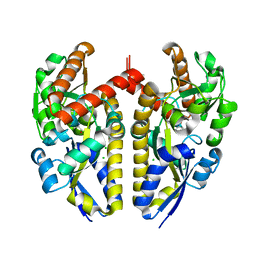 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol. | | Descriptor: | (2S)-2-aminopropan-1-ol, CHLORIDE ION, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol.
To be published
|
|
4XFR
 
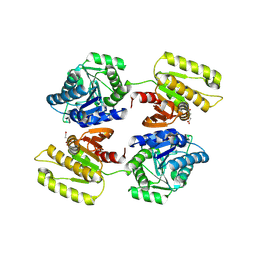 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group P6522 | | Descriptor: | CITRIC ACID, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-28 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XGJ
 
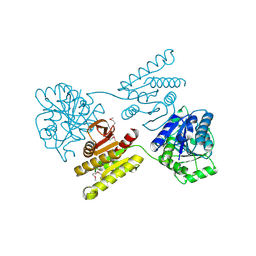 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, APO structure, domain swapped dimer | | Descriptor: | Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-02-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XDG
 
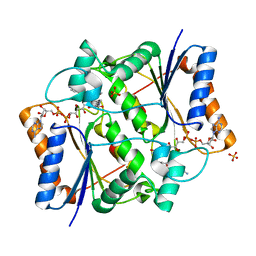 | | Crystal Structure of Quinone Reductase II in complex with 2-(4-aminophenyl)-5-methoxy-1-oxy-indol-3-one molecule | | Descriptor: | (2S)-2-(4-aminophenyl)-1-hydroxy-5-methoxy-1,2-dihydro-3H-indol-3-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Sirigu, S, Nepveu, F, Vuillard, L, Ferry, G, Isabet, T, Thompson, A, Boutin, J.A. | | Deposit date: | 2014-12-19 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Quinone Reductase 2 in the Antimalarial Properties of Indolone-Type Derivatives.
Molecules, 22, 2017
|
|
4XFM
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, with bound D-threonate, domain swapped dimer | | Descriptor: | THREONATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-27 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XIY
 
 | | Crystal structure of ketol-acid reductoisomerase from Azotobacter | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Ketol-acid reductoisomerase, ... | | Authors: | Spatzal, T, Cahn, J.K.B, Wiig, J.A, Einsle, O, Hu, Y, Ribbe, M.W, Arnold, F.H. | | Deposit date: | 2015-01-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
4XDH
 
 | | Crystal Structure of Quinone Reductase II in complex with a 2-(4-methoxy-phenyl)-5-methoxy-indol-3-one molecule | | Descriptor: | 5-methoxy-2-(4-methoxyphenyl)-3H-indol-3-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Sirigu, S, Nepveu, F, Vuillard, L, Ferry, G, Isabet, T, Thompson, A, Boutin, J.A. | | Deposit date: | 2014-12-19 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Quinone Reductase 2 in the Antimalarial Properties of Indolone-Type Derivatives.
Molecules, 22, 2017
|
|
4XFE
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, SULFATE ION, TRAP dicarboxylate transporter subunit DctP, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate
To be published
|
|
4XG0
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group C2221 | | Descriptor: | CHLORIDE ION, CITRIC ACID, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XZQ
 
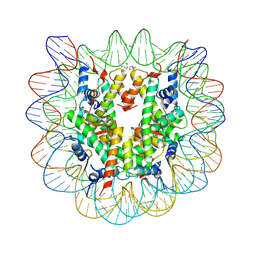 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
4XUF
 
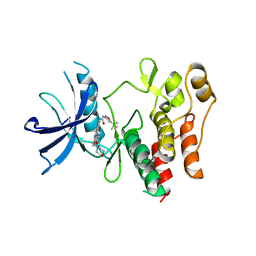 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor quizartinib (AC220) | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zorn, J.A, Wang, Q, Fujimura, E, Barros, T, Kuriyan, J. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the FLT3 Kinase Domain Bound to the Inhibitor Quizartinib (AC220).
Plos One, 10, 2015
|
|
4XX9
 
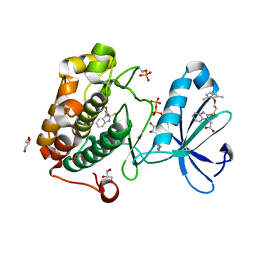 | |
4XZ0
 
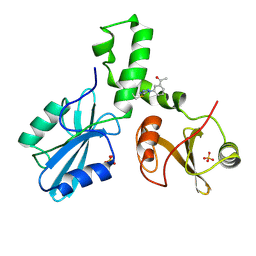 | | ZAP-70-tSH2:compound-A complex | | Descriptor: | 1-(3-{5-[(3-chlorobenzyl)sulfonyl]-1H-tetrazol-1-yl}phenyl)ethanone, SULFATE ION, Tyrosine-protein kinase ZAP-70 | | Authors: | Barros, T, Kuriyan, J, Visperas, P.R, Winger, J.A. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modification by covalent reaction or oxidation of cysteine residues in the tandem-SH2 domains of ZAP-70 and Syk can block phosphopeptide binding.
Biochem. J., 465, 2015
|
|
4XZS
 
 | | Crystal Structure of TRIAP1-MBP fusion | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Abid-Ali, F, Perez-Dorado, I, Matthews, S.J. | | Deposit date: | 2015-02-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
4XZ1
 
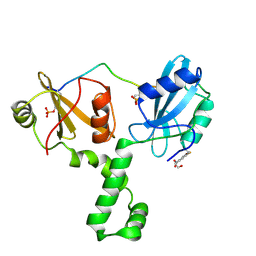 | | ZAP-70-tSH2:Compound-B adduct | | Descriptor: | 2-[(7-chloro-4-nitro-2,1,3-benzoxadiazol-5-yl)amino]ethanol, Tyrosine-protein kinase ZAP-70, doubly phosphorylated ITAM peptide | | Authors: | Barros, T, Kuriyan, J, Winger, J.A, Visperas, P.R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modification by covalent reaction or oxidation of cysteine residues in the tandem-SH2 domains of ZAP-70 and Syk can block phosphopeptide binding.
Biochem. J., 465, 2015
|
|
4XZV
 
 | | Crystal Structure of SLMO1-TRIAP1 Complex | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, Protein slowmo homolog 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
4YKQ
 
 | | Heat Shock Protein 90 Bound to CS301 | | Descriptor: | 1-(5-ethyl-2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS301
To Be Published
|
|
4YBK
 
 | | C-Helix-Out Dasatinib Analog Crystallized with c-Src Kinase | | Descriptor: | 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(4-phenoxyphenyl)-1,3-thiazole-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.K, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
4YEX
 
 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
