5DZT
 
 | |
6UAK
 
 | | LahSb - C-terminal methyltransferase involved in RiPP biosynthesis | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM dependent methyltransferase LahSB | | Authors: | Nair, S.K, Estrada, P. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of a Dehydratase and Methyltransferase in the Biosynthesis of Ribosomally Synthesized and Post-translationally Modified Peptides in Lachnospiraceae.
Chembiochem, 21, 2020
|
|
6C0Y
 
 | |
6EC7
 
 | |
6EC8
 
 | |
5U55
 
 | | Psf4 in complex with Mn2+ and (S)-2-HPP | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5U5D
 
 | | Psf4 in complex with Mn2+ and (R)-2-HPP | | Descriptor: | (S)-2-hydroxypropylphosphonic acid epoxidase, MANGANESE (II) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
7MSK
 
 | | ThuS glycosin S-glycosyltransferase | | Descriptor: | Glyco_trans_2-like domain-containing protein, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSN
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSP
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | MAGNESIUM ION, SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
6M7Y
 
 | |
6MPZ
 
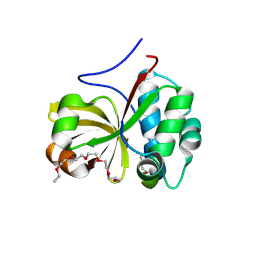 | | Crystal structure of a double glycine motif protease from AMS/PCAT transporter in complex with the leader peptide | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Double Glycine Motif Protease domain from AMS/PCAT Transporter, peptide aldehyde inhibitor 1 based on the ProcA2.8 leader peptide | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2018-10-09 | | Release date: | 2019-02-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease.
Elife, 8, 2019
|
|
6B9T
 
 | |
6B9S
 
 | |
6C0H
 
 | |
6B9R
 
 | |
6C0G
 
 | |
5U58
 
 | | Psf4 in complex with Fe2+ and (R)-2-HPP | | Descriptor: | (S)-2-hydroxypropylphosphonic acid epoxidase, FE (III) ION, [(2R)-2-hydroxypropyl]phosphonic acid | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5U57
 
 | | Psf4 in complex with Fe2+ and (S)-2-HPP | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, (S)-2-hydroxypropylphosphonic acid epoxidase, FE (III) ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5U5G
 
 | | Psf3 in complex with NADP+ and 2-OPP | | Descriptor: | (2-oxopropyl)phosphonic acid, 6-phosphogluconate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olivares, P, Nair, S.K. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.
ACS Chem. Biol., 12, 2017
|
|
5WA4
 
 | |
5W98
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PbtD | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W99
 
 | | Pyridine synthase, PbtD, from GE2270 biosynthesis bound to TSP | | Descriptor: | 2,2'-(6-(2'-(aminomethyl)-[2,4'-bithiazol]-4-yl)pyridine-2,5-diyl)bis(thiazole-4-carboxylic acid), PbtD, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2017-06-22 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WA3
 
 | |
5UAO
 
 | | Crystal structure of MibH, a lathipeptide tryptophan 5-halogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophane-5-halogenase | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Two Flavoenzymes Catalyze the Post-Translational Generation of 5-Chlorotryptophan and 2-Aminovinyl-Cysteine during NAI-107 Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
