6PYB
 
 | | Sex Hormone-binding globulin mutant E176K in complex with DVT | | Descriptor: | 4,4'-[(3R,4R)-oxolane-3,4-diylbis(methylene)]bis(2-methoxyphenol), CALCIUM ION, Sex hormone-binding globulin | | Authors: | Round, P.W, Das, S, Van Petegem, F. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular interactions between sex hormone-binding globulin and nonsteroidal ligands that enhance androgen activity.
J.Biol.Chem., 295, 2020
|
|
7TCI
 
 | | Structure of Xenopus KCNQ1-CaM in complex with ML277 | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, CALCIUM ION, Calmodulin-1, ... | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
7TCP
 
 | | Structure of Xenopus KCNQ1-CaM | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Willegems, K, Kyriakis, E, Van Petegem, F, Eldstrom, J, Fedida, D. | | Deposit date: | 2021-12-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277.
Nat Commun, 13, 2022
|
|
7CF9
 
 | | Structure of RyR1 (Ca2+/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6MUD
 
 | |
6MUE
 
 | |
6M2W
 
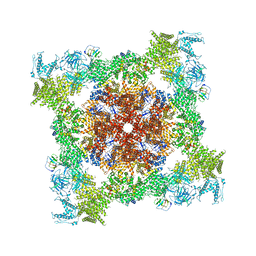 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
9BMK
 
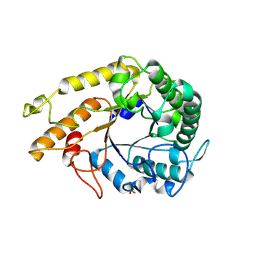 | | GH5_4 endo-beta(1,3/1,4)-glucanase E331A from Segatella copri in complex with cellotriose | | Descriptor: | Cellulase (Glycosyl hydrolase family 5), SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cordeiro, R.L, Golisch, B, Brumer, H, Van Petegem, F. | | Deposit date: | 2024-05-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The molecular basis of cereal mixed-linkage beta-glucan utilization by the human gut bacterium Segatella copri.
J.Biol.Chem., 300, 2024
|
|
9BAM
 
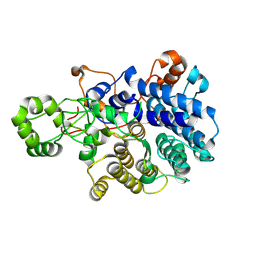 | | Surface glycan-binding protein A (SGBP-A, SusD-like) from a mixed-linkage beta-glucan utilization locus in Segatella copri | | Descriptor: | MAGNESIUM ION, Surface glycan-binding protein A (SGBP-A) | | Authors: | Cordeiro, R.L, Golisch, B, Brumer, H, Van Petegem, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The molecular basis of cereal mixed-linkage beta-glucan utilization by the human gut bacterium Segatella copri.
J.Biol.Chem., 300, 2024
|
|
9BJX
 
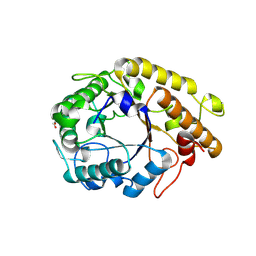 | | GH5_4 endo-beta(1,3/1,4)-glucanase from Segatella copri | | Descriptor: | 1,2-ETHANEDIOL, Cellulase (Glycosyl hydrolase family 5) | | Authors: | Cordeiro, R.L, Golisch, B, Brumer, H, Van Petegem, F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular basis of cereal mixed-linkage beta-glucan utilization by the human gut bacterium Segatella copri.
J.Biol.Chem., 300, 2024
|
|
9BAL
 
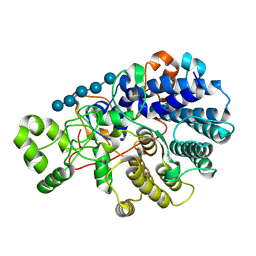 | | Surface glycan-binding protein A (SGBP-A, SusD-like) from a mixed-linkage beta-glucan utilization locus in Segatella copri in complex with cellopentaose | | Descriptor: | MAGNESIUM ION, Surface glycan-binding protein A (SGBP-A), beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cordeiro, R.L, Golisch, B, Brumer, H, Van Petegem, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of cereal mixed-linkage beta-glucan utilization by the human gut bacterium Segatella copri.
J.Biol.Chem., 300, 2024
|
|
6PAL
 
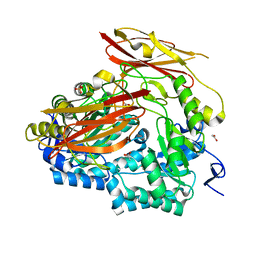 | | Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus | | Descriptor: | ACETATE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tamura, K, Brumer, H, van Petegem, F. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human GutBacteroides.
Mbio, 11, 2020
|
|
8F5B
 
 | | Human ABCA4 structure in complex with AMP-PNP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Scortecci, J.F, Van Petegem, F, Molday, R.S. | | Deposit date: | 2022-11-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural and functional characterization of the nucleotide-binding domains of ABCA4 and their role in Stargardt disease.
J.Biol.Chem., 300, 2024
|
|
1PG5
 
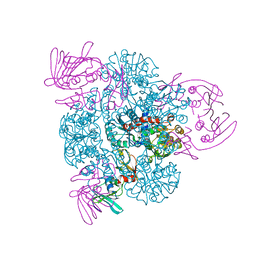 | | CRYSTAL STRUCTURE OF THE UNLIGATED (T-STATE) ASPARTATE TRANSCARBAMOYLASE FROM THE EXTREMELY THERMOPHILIC ARCHAEON SULFOLOBUS ACIDOCALDARIUS | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | De Vos, D, Van Petegem, F, Remaut, H, Legrain, C, Glansdorff, N, Van Beeumen, J.J. | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of T State Aspartate Carbamoyltransferase of the Hyperthermophilic Archaeon Sulfolobus acidocaldarius.
J.Mol.Biol., 339, 2004
|
|
9C1E
 
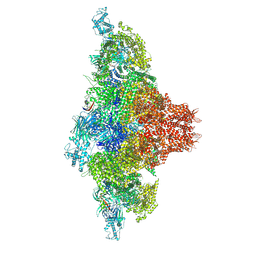 | | Mink RyR3 in closed conformation | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 3, ... | | Authors: | Chen, Y.S, Van Petegem, F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM investigation of ryanodine receptor type 3.
Nat Commun, 15, 2024
|
|
9C1F
 
 | |
5SV8
 
 | | Crystal Structure of the catalytic nucleophile and surface cysteine mutant of VvEG16 in complex with a xyloglucan oligosaccharide | | Descriptor: | alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, probable xyloglucan endotransglucosylase/hydrolase protein 19 | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
5VSN
 
 | |
5FDY
 
 | |
5FEB
 
 | |
5C33
 
 | |
5C30
 
 | |
8SEP
 
 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEU
 
 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | Descriptor: | Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
