6LK7
 
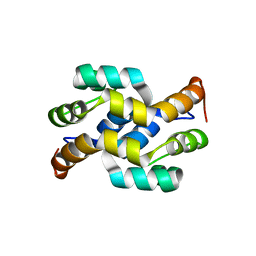 | | crystal structure of Os1348 from Pseudomonas sp. Os17 | | Descriptor: | Nitrile hydratase, alpha chain | | Authors: | Takeuchi, K, Tsuchiya, W, Fujimoto, Z, Yamada, K, Someya, N, Yamazaki, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Discovery of an Antibiotic-Related Small Protein of Biocontrol Strain Pseudomonas sp. Os17 by a Genome-Mining Strategy.
Front Microbiol, 11, 2020
|
|
3RJ1
 
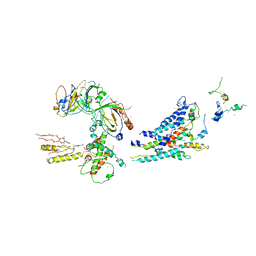 | | Architecture of the Mediator Head module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Imasaki, T, Calero, G, Cai, G, Tsai, K.L, Yamada, K, Cardelli, F, Erdjument-Bromage, H, Tempst, P, Berger, I, Kornberg, G.L, Asturias, F.J, Kornberg, R.D, Takagi, Y. | | Deposit date: | 2011-04-14 | | Release date: | 2011-07-27 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Architecture of the Mediator head module.
Nature, 475, 2011
|
|
4JGT
 
 | | Structure and kinetic analysis of H2S production by human Mercaptopyruvate Sulfurtransferase | | Descriptor: | 3-mercaptopyruvate sulfurtransferase, GLYCEROL, PYRUVIC ACID, ... | | Authors: | Koutmos, M, Yamada, K, Yadav, P.K, Chiku, T, Banerjee, R. | | Deposit date: | 2013-03-03 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Structure and Kinetic Analysis of H2S Production by Human Mercaptopyruvate Sulfurtransferase.
J.Biol.Chem., 288, 2013
|
|
5UJC
 
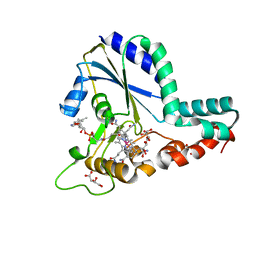 | | Crystal structure of a C.elegans B12-trafficking protein CblC, a human MMACHC homologue | | Descriptor: | CO-METHYLCOBALAMIN, D(-)-TARTARIC ACID, GLYCEROL, ... | | Authors: | Li, Z, Shanmuganathan, A, Ruetz, M, Yamada, K, Lesniak, N.A, Krautler, B, Brunold, T.C, Banerjee, R, Koutmos, M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Coordination chemistry controls the thiol oxidase activity of the B12-trafficking protein CblC.
J. Biol. Chem., 292, 2017
|
|
1WUA
 
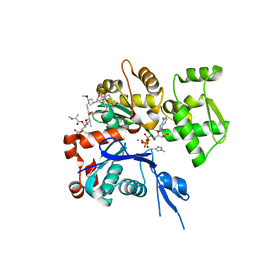 | | The structure of Aplyronine A-actin complex | | Descriptor: | (8R,9R,10R,11R,14S,18S,20S,24S)-24-{(1R,2S,3R,6R,7R,8R,9S,10E)-8-(ACETYLOXY)-6-[(N,N-DIMETHYLALANYL)OXY]-11-[FORMYL(MET HYL)AMINO]-2-HYDROXY-1,3,7,9-TETRAMETHYLUNDEC-10-ENYL}-10-HYDROXY-14,20-DIMETHOXY-9,11,15,18-TETRAMETHYL-2-OXOOXACYCLOTE TRACOSA-3,5,15,21-TETRAEN-8-YL N,N,O-TRIMETHYLSERINATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Hirata, K, Muraoka, S, Suenaga, K, Kuroda, T, Kato, K, Tanaka, H, Yamamoto, M, Takata, M, Yamada, K, Kigoshi, H. | | Deposit date: | 2004-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure basis for antitumor effect of aplyronine a
J.Mol.Biol., 356, 2006
|
|
4YTK
 
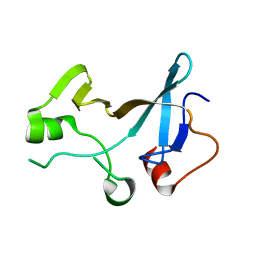 | | Structure of the KOW1-Linker1 domain of Transcription Elongation Factor Spt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, Zhang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.0904 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4YTL
 
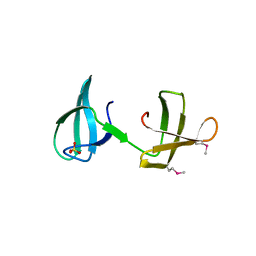 | | Structure of the KOW2-KOW3 Domain of Transcription Elongation Factor Spt5. | | Descriptor: | GLYCEROL, SULFATE ION, Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, ZHang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
7EFV
 
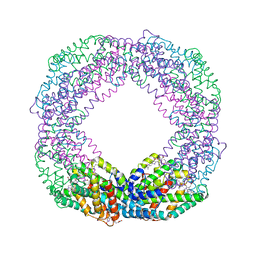 | | Crystal structure of octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Hung, N.K, Yamada, K, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EH7
 
 | | Cryo-EM structure of the octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EFW
 
 | | Crystal structure of hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCEROL, ... | | Authors: | Minato, T, Teramoto, T, Hung, N.K, Yamada, K, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EH8
 
 | | Cryo-EM structure of the hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
5WDY
 
 | |
5GUA
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138Y mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU9
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138I mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5DRB
 
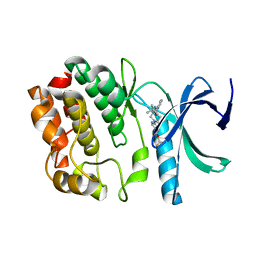 | | Crystal structure of WNK1 in complex with WNK463 | | Descriptor: | N-tert-butyl-1-(1-{5-[5-(trifluoromethyl)-1,3,4-oxadiazol-2-yl]pyridin-2-yl}piperidin-4-yl)-1H-imidazole-5-carboxamide, Serine/threonine-protein kinase WNK1 | | Authors: | Kohls, D, Xie, X. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Small-molecule WNK inhibition regulates cardiovascular and renal function.
Nat.Chem.Biol., 12, 2016
|
|
5WE8
 
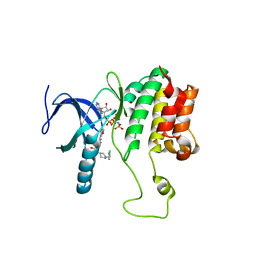 | |
5TF9
 
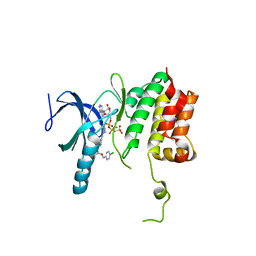 | |
8RKI
 
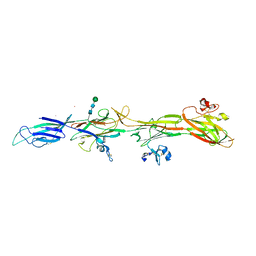 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8BQU
 
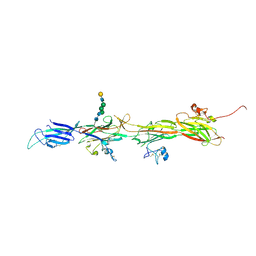 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8W86
 
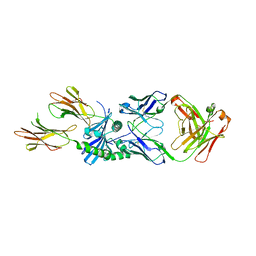 | | HLA-DQ2.5-B/C hordein peptide in complex with DQN0385AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE02 Fab heavy chain, DQN0385AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W85
 
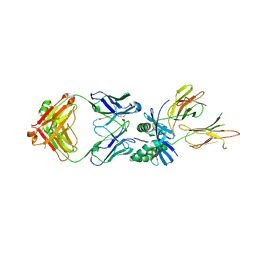 | | HLA-DQ2.5-gamma2 gliadin peptide in complex with DQN0385AE01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE01 Fab heavy chain, DQN0385AE01 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W84
 
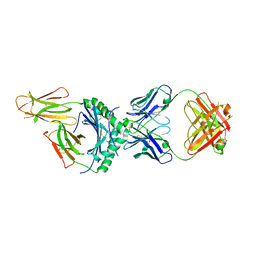 | | HLA-DQ2.5-alpha2 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W83
 
 | | HLA-DQ2.5-alpha1 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
3LJU
 
 | | Crystal structure of full length centaurin alpha-1 bound with the head group of PIP3 | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, Arf-GAP with dual PH domain-containing protein 1, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
