1S3N
 
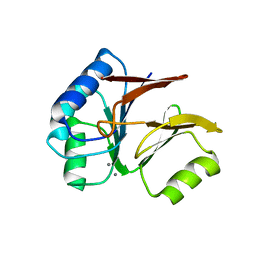 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, MANGANESE (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1S3L
 
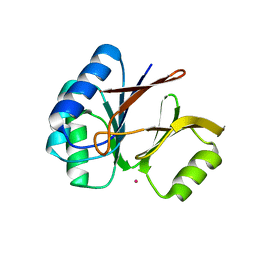 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4JHC
 
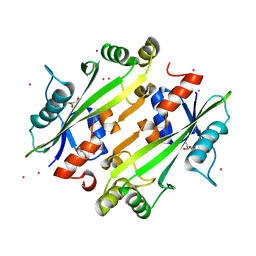 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli | | Descriptor: | GLYCEROL, Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2013-03-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
4LU1
 
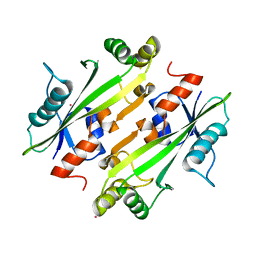 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli, mutant D69A | | Descriptor: | Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
3CBT
 
 | | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor | | Descriptor: | MAGNESIUM ION, Phosphatase SC4828, SODIUM ION | | Authors: | Singer, A.U, Xu, X, Chang, C, Zheng, H, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor.
To be Published
|
|
3D1R
 
 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3DRW
 
 | | Crystal Structure of a Phosphofructokinase from Pyrococcus horikoshii OT3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-specific phosphofructokinase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Kochinyan, S, Brown, G, Cuff, M.E, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Jia, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
3EXM
 
 | | Crystal structure of the phosphatase SC4828 with the non-hydrolyzable nucleotide GPCP | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID GUANOSYL ESTER, ... | | Authors: | Singer, A.U, Xu, X, Zheng, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of a new family of prokaryotic nucleoside diphosphatases.
To be Published
|
|
3EXC
 
 | | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Singer, A.U, Skarina, T, Tan, K, Kagan, O, Onopriyenko, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus
To be Published
|
|
3FHM
 
 | | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Zhang, R, Cui, H, Kudritsdka, M, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
To be Published
|
|
3F4F
 
 | | Crystal structure of dUT1p, a dUTPase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
4Q92
 
 | | 1.90 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-modified Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QN2
 
 | | 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with NAD+ and BME-free Cys289 | | Descriptor: | ACETATE ION, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QJE
 
 | | 1.85 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-free sulfinic acid form of Cys289 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3OI7
 
 | | Structure of the structure of the H13A mutant of Ykr043C in complex with sedoheptulose-1,7-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,7-di-O-phosphono-beta-D-altro-hept-2-ulofuranose, GLYCEROL, ... | | Authors: | Singer, A.U, Xu, X, Dong, A, Cui, H, Clasquin, M.F, Caudy, A.A, Edwards, A.M, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Riboneogenesis in yeast.
Cell(Cambridge,Mass.), 145, 2011
|
|
3Q3C
 
 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3PT1
 
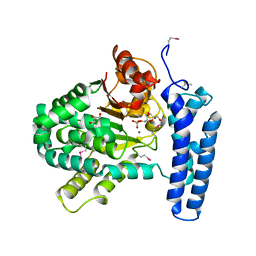 | | Structure of DUF89 from Saccharomyces cerevisiae co-crystallized with F6P. | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure and activity of a DUF89 protein from Saccharomyces cerevisiae revealed a novel family of carbohydrate phosphatases
To be Published
|
|
3R3U
 
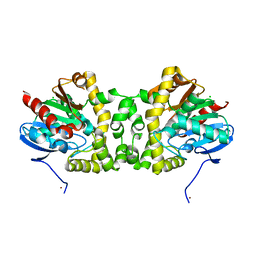 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, NICKEL (II) ION | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R41
 
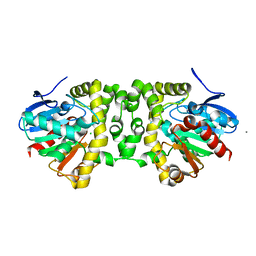 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/apo | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-17 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3QYJ
 
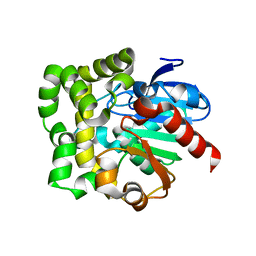 | |
3QYF
 
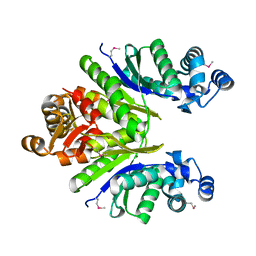 | | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus | | Descriptor: | CRISPR-ASSOCIATED PROTEIN, MAGNESIUM ION | | Authors: | Petit, P, Xu, X, Chang, C, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus
To be Published
|
|
3R3V
 
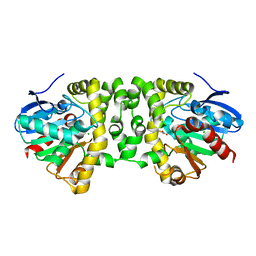 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Fluoroacetate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase, ... | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R0P
 
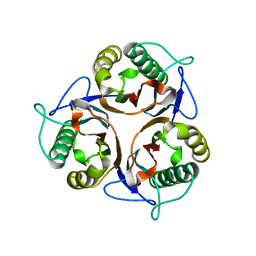 | | Crystal structure of L-PSP putative endoribonuclease from uncultured organism | | Descriptor: | L-PSP putative endoribonuclease | | Authors: | Cuff, M.E, Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-PSP putative endoribonuclease from uncultured organism
To be Published
|
|
