3KK0
 
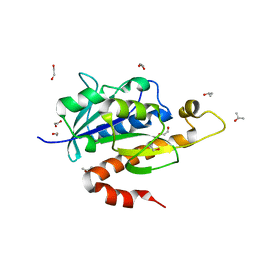 | | Crystal structure of partially folded intermediate state of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Singh, N, Yadav, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Fully-Folded Native and Partially-Folded Intermediate States of Peptidyl-tRNA Hydrolase from Mycobacterium smegmatis
To be Published
|
|
5JXX
 
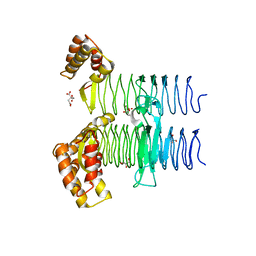 | | Crystal structure of UDP-N-acetylglucosamine O-acyltransferase (LpxA) from Moraxella catarrhalis RH4. | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION, GLYCEROL | | Authors: | Pratap, S, Kesari, P, Yadav, R, Narwal, M, Dev, A, Kumar, P. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Acyl chain preference and inhibitor identification of Moraxella catarrhalis LpxA: Insight through crystal structure and computational studies.
Int. J. Biol. Macromol., 96, 2017
|
|
3P2J
 
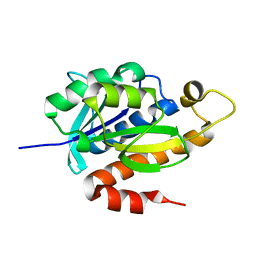 | | Crystal structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Yadav, R, Sinha, M, Arora, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution
To be Published
|
|
4ZXP
 
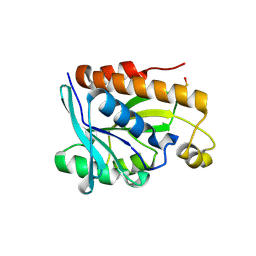 | | Crystal structure of Peptidyl- tRNA Hydrolase from Vibrio cholerae | | Descriptor: | CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Shahid, S, Pal, R.K, Kabra, A, Yadav, R, Kumar, A, Arora, A. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.
RNA, 23, 2017
|
|
5GO2
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Citrate | | Descriptor: | CITRIC ACID, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
5YOZ
 
 | |
5GMU
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
8DQF
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | Descriptor: | Carbonic anhydrase, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DYQ
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DR2
 
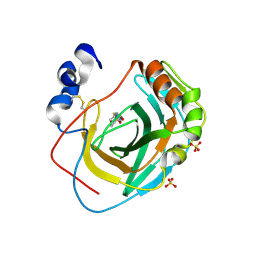 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide | | Descriptor: | 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DRB
 
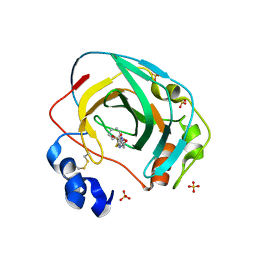 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 3-phenyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide | | Descriptor: | 3-phenyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DPC
 
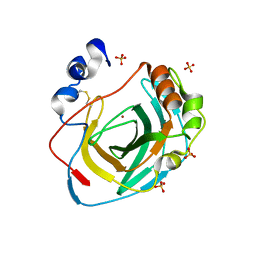 | | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae | | Descriptor: | Carbonic anhydrase, SULFATE ION, ZINC ION | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
2LXX
 
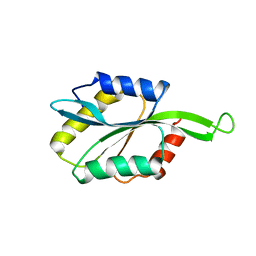 | | Solution structure of cofilin like UNC-60B protein from Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 2, isoform c | | Authors: | Shukla, V, Yadav, R, Kabra, A, Jain, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of UNC-60B from Caenorhabditis elegans
To be Published
|
|
2MJL
 
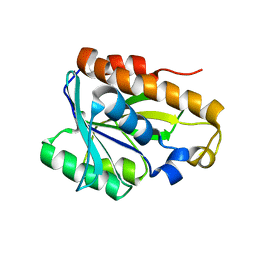 | | Solution structure of peptidyl-tRNA hyrolase from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kabra, A, Shahid, S, Yadav, R, Pulavarti, S, Shukla, V.K, Arora, A. | | Deposit date: | 2014-01-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptidyl-tRNA hydrolase from Vibrio cholerae
To be Published
|
|
2MP4
 
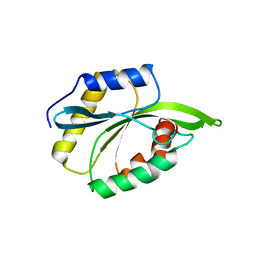 | | Solution Structure of ADF like UNC-60A Protein of Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 1, isoforms a/b | | Authors: | Shukla, V, Yadav, R, Kabra, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone dynamics of ADF like UNC-60A protein from Caenorhabditis elegans: its divergence from conventional ADF/cofilin
To be Published
|
|
2LRA
 
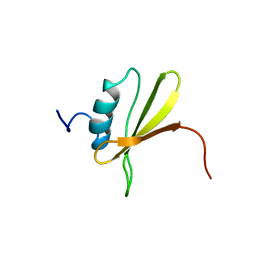 | | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S, Yadav, R, Jain, A, Pathak, P, Meher, A, Arora, A. | | Deposit date: | 2012-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag
To be Published
|
|
5XCJ
 
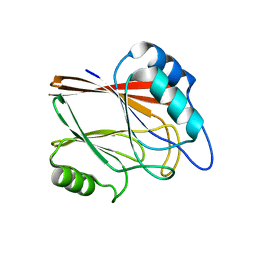 | | Crystal structure of Vps29 single mutant (D62A) from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
8HPT
 
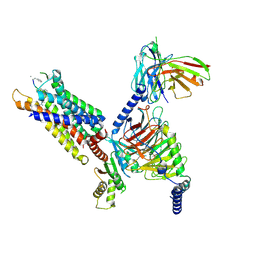 | | Structure of C5a-pep bound mouse C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saha, S, Maharana, J, Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8HQC
 
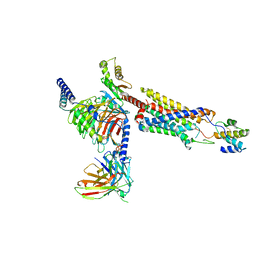 | | Structure of a GPCR-G protein in complex with a natural peptide agonist | | Descriptor: | Antibody fragment, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Saha, S, Maharana, J, Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
5B6J
 
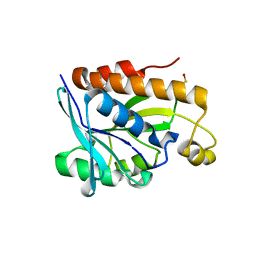 | |
6XM2
 
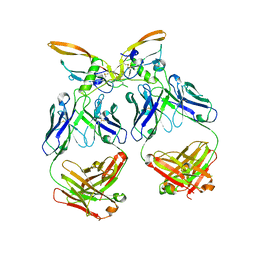 | | The structure of the 4A11.v7 antibody in complex with human TGFb2 | | Descriptor: | 4A11.v7 heavy chain Fab (VH-CH1) IgG1 humanized, 4A11.v7 kappa light chain Fab (VL-CL) humanized, Transforming growth factor beta-2 | | Authors: | Lupardus, P.J, Yin, J.P. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | TGF beta 2 and TGF beta 3 isoforms drive fibrotic disease pathogenesis.
Sci Transl Med, 13, 2021
|
|
6CIR
 
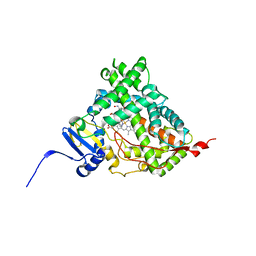 | | Human Cytochrome P450 17A1 in complex with inhibitor: abiraterone C6 oxime | | Descriptor: | 6-oxime-17-(3-pyridyl)-androst-16-en-3-ol, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E, Fehl, C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structure-Based Design of Inhibitors with Improved Selectivity for Steroidogenic Cytochrome P450 17A1 over Cytochrome P450 21A2.
J. Med. Chem., 61, 2018
|
|
6CIZ
 
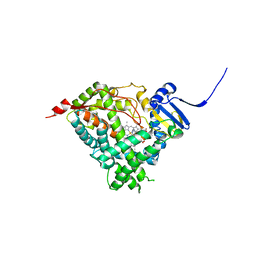 | | Human Cytochrome P450 17A1 in complex with inhibitor: abiraterone C6 nitrile | | Descriptor: | 6-cyano-17-(3-pyridyl)-androst-5,16-dien-3-ol, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E, Fehl, C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure-Based Design of Inhibitors with Improved Selectivity for Steroidogenic Cytochrome P450 17A1 over Cytochrome P450 21A2.
J. Med. Chem., 61, 2018
|
|
6CHI
 
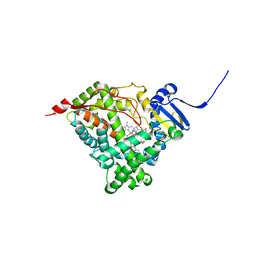 | | Human Cytochrome P450 17A1 in complex with inhibitor: abiraterone C6 amide | | Descriptor: | 3-hydroxy-17-(3-pyridyl)-androst-5,16-dien-6-amide, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structure-Based Design of Inhibitors with Improved Selectivity for Steroidogenic Cytochrome P450 17A1 over Cytochrome P450 21A2.
J. Med. Chem., 61, 2018
|
|
4Z86
 
 | |
