6J7N
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7S
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) wild type protein from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7P
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant E146Q co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.629 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7Q
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A from Mycobacterium tuberculosis | | Descriptor: | CALCIUM ION, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
7XT3
 
 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
6M40
 
 | | Crystal structure of the NS3-like helicase from Alongshan virus | | Descriptor: | NS3-like protein | | Authors: | Gao, X.P, Zhu, K.X, Chen, P, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2020-03-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the NS3-like helicase from Alongshan virus.
Iucrj, 7, 2020
|
|
5KWB
 
 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
3TIP
 
 | |
3TIQ
 
 | | Crystal structure of Staphylococcus aureus SasG G51-E-G52 module | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Surface protein G | | Authors: | Gruszka, D.T, Wojdyla, J.A, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8739 Å) | | Cite: | Staphylococcal biofilm-forming protein has a contiguous rod-like structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GZF
 
 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5EW5
 
 | |
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5GNB
 
 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5WWP
 
 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
5XGQ
 
 | |
5XET
 
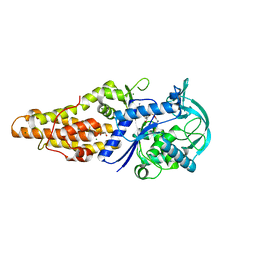 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
7CMD
 
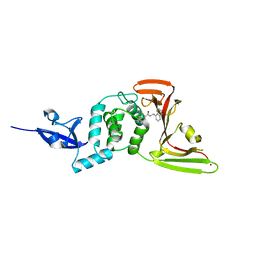 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CJD
 
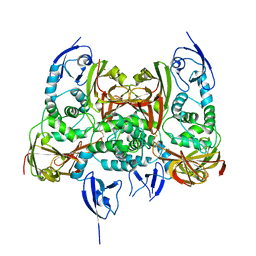 | |
6YWK
 
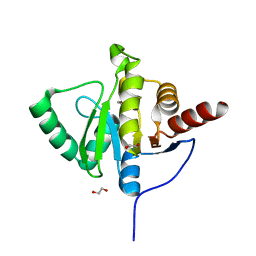 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YWM
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6T0O
 
 | | Crystal structure of YTHDC1 with fragment 14 (ACA_DC1_004) | | Descriptor: | 2-methyl-3~{H}-pyrido[3,4-d]pyrimidin-4-one, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZT
 
 | | Crystal structure of YTHDC1 with fragment 10 (DHU_DC1_076) | | Descriptor: | 6-[[methyl(thiophen-3-ylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T02
 
 | | Crystal structure of YTHDC1 with fragment 15 (DHU_DC1_169) | | Descriptor: | (~{S})-phenyl-[(2~{S})-pyrrolidin-2-yl]methanol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
