5IJ4
 
 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | Descriptor: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | Authors: | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
1Z9E
 
 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | Descriptor: | PC4 and SFRS1 interacting protein 2 | | Authors: | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | Deposit date: | 2005-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
1CSA
 
 | |
1CDB
 
 | |
2GUT
 
 | | Solution structure of the trans-activation domain of the human co-activator ARC105 | | Descriptor: | ARC/MEDIATOR, Positive cofactor 2 glutamine/Q-rich-associated protein | | Authors: | Vought, B.W, Jim Sun, Z.-Y, Hyberts, S.G, Wagner, G, Naar, A.M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis.
Nature, 442, 2006
|
|
2ONT
 
 | | A swapped dimer of the HIV-1 capsid C-terminal domain | | Descriptor: | Capsid protein p24 | | Authors: | Ivanov, D, Tsodikov, O.V, Kasanov, J, Ellenberger, T, Wagner, G, Collins, T. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Domain-swapped dimerization of the HIV-1 capsid C-terminal domain
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7L0R
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2OGH
 
 | | Solution structure of yeast eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Reibarkh, M, del Rio, F, Yamamoto, Y, Asano, K, Wagner, G. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Initiation Factor (eIF) 1 Carries Two Distinct eIF5-binding Faces Important for Multifactor Assembly and AUG Selection.
J.Biol.Chem., 283, 2008
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2RG8
 
 | |
2ROQ
 
 | | Solution Structure of the thiolation-thioesterase di-domain of enterobactin synthetase component F | | Descriptor: | Enterobactin synthetase component F | | Authors: | Frueh, D.P, Arthanari, H, Koglin, A, Vosburg, D.A, Bennett, A.E, Walsh, C.T, Wagner, G. | | Deposit date: | 2008-04-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase
Nature, 454, 2008
|
|
2PV6
 
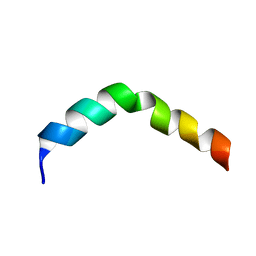 | | HIV-1 gp41 Membrane Proximal Ectodomain Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein | | Authors: | Sun, Z.-Y.J, Oh, K.J, Kim, M, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-05-09 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane
Immunity, 28, 2008
|
|
2RON
 
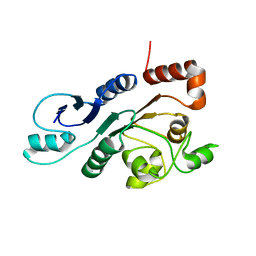 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
1NFA
 
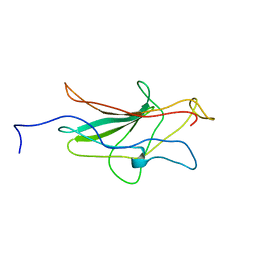 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | Authors: | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
2B86
 
 | |
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
2BID
 
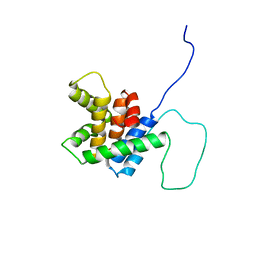 | | HUMAN PRO-APOPTOTIC PROTEIN BID | | Descriptor: | PROTEIN (BID) | | Authors: | Chou, J.J, Li, H, Salvesen, G.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-01-27 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BID, an intracellular amplifier of apoptotic signaling.
Cell(Cambridge,Mass.), 96, 1999
|
|
1ZGW
 
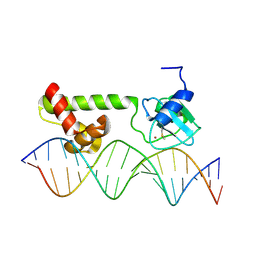 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1NUI
 
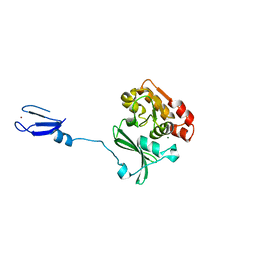 | | Crystal Structure of the primase fragment of Bacteriophage T7 primase-helicase protein | | Descriptor: | DNA primase/helicase, MAGNESIUM ION, ZINC ION | | Authors: | Kato, M, Ito, T, Wagner, G, Richardson, C.C, Ellenberger, T. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modular Architecture of the Bacteriophage T7 Primase Couples RNA primer Synthesis to DNA Synthesis
Mol.Cell, 11, 2003
|
|
1L3E
 
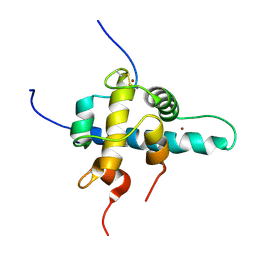 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1P4Q
 
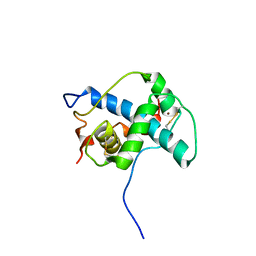 | | Solution structure of the CITED2 transactivation domain in complex with the p300 CH1 domain | | Descriptor: | Cbp/p300-interacting transactivator 2, E1A-associated protein p300, ZINC ION | | Authors: | Freedman, S.J, Sun, Z.-Y.J, Kung, A.L, France, D.S, Wagner, G, Eck, M.J. | | Deposit date: | 2003-04-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2.
Nat.Struct.Biol., 10, 2003
|
|
1N4Y
 
 | | REFINED STRUCTURE OF KISTRIN | | Descriptor: | KISTRIN | | Authors: | Krezel, A.M, Krane, J, Dennis, M.S, Lazarus, R.A, Wagner, G. | | Deposit date: | 2002-11-02 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist.
Science, 253, 1991
|
|
2AIV
 
 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
