1ACJ
 
 | |
2ACK
 
 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, MONOCHROMATIC DATA | | Descriptor: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | Authors: | Raves, M.L, Sussman, J.L, Harel, M, Silman, I. | | Deposit date: | 1997-10-29 | | Release date: | 1998-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
8D35
 
 | |
1ODC
 
 | | STRUCTURE OF ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH N-4'-QUINOLYL-N'-9"-(1",2",3",4"-TETRAHYDROACRIDINYL)-1,8- DIAMINOOCTANE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N-QUINOLIN-4-YL-N'-(1,2,3,4-TETRAHYDROACRIDIN-9-YL)OCTANE-1,8-DIAMINE | | Authors: | Wong, D.M, Greenblatt, H.M, Carlier, P.R, Han, Y.-F, Pang, Y.-P, Silman, I, Sussman, J.L. | | Deposit date: | 2003-02-15 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complexes of Alkylene-Linked Tacrine Dimers with Torpedo Californica Acetylcholinesterase: Binding of Bis(5)-Tacrine Produces a Dramatic Rearrangement in the Active-Site Gorge.
J.Med.Chem., 49, 2006
|
|
1UT6
 
 | | Structure of acetylcholinesterase (E.C. 3.1.1.7) complexed with N-9-(1',2',3',4'-Tetrahydroacridinyl)-1,8- diaminooctane at 2.4 angstroms resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N-9-(1',2',3',4'-TETRAHYDROACRIDINYL)-1,8-DIAMINOOCTANE | | Authors: | Brumshtein, B, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.-P, Silman, I, Sussman, J.L. | | Deposit date: | 2003-12-04 | | Release date: | 2005-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complexes of Alkylene-Linked Tacrine Dimers with Torpedo Californica Acetylcholinesterase: Binding of Bis(5)-Tacrine Produces a Dramatic Rearrangement in the Active-Site Gorge.
J.Med.Chem., 49, 2006
|
|
3G9V
 
 | | Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22 | | Descriptor: | Interleukin 22 receptor, alpha 2, Interleukin-22 | | Authors: | de Moura, P.R, Watanabe, L, Bleicher, L, Colau, D, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2009-02-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22
FEBS Lett., 583, 2009
|
|
7OX5
 
 | | hIL-9:hIL-9Ra complex | | Descriptor: | Interleukin-9, Interleukin-9 receptor, SULFATE ION | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7OX6
 
 | | Solution structure of human interleukin-9 | | Descriptor: | Interleukin-9 | | Authors: | Savvides, S.N, Tripsianes, K, De Vos, T, Papageorgiou, A, Evangelidis, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-11 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7OX4
 
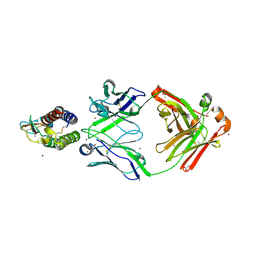 | | Mouse interleukin-9 in complex with Fab 35D8. | | Descriptor: | ACETATE ION, Heavy chain (Fab 35D8), Interleukin-9, ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7OX3
 
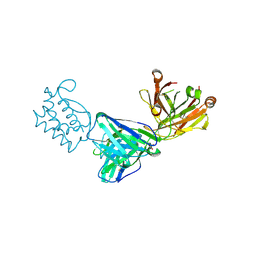 | | Fab 6D3: hIL-9 complex | | Descriptor: | Heavy chain (Fab 6D3), Interleukin-9, Light chain (Fab 6D3), ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7OX2
 
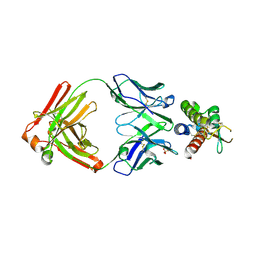 | | Fab 6E2: hIL-9 complex | | Descriptor: | Heavy chain (Fab 6E2), Interleukin-9, Light chain (Fab 6E2), ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7OX1
 
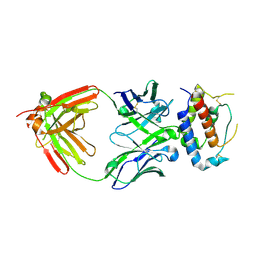 | | Fab 7D6: hIL-9 complex | | Descriptor: | Heavy chain (Fab 7D6), Interleukin-9, Light chain (Fab 7D6) | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
3N3K
 
 | | The catalytic domain of USP8 in complex with a USP8 specific inhibitor | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 8, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Allali-Hassani, A, Lam, R, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3MTN
 
 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
6G0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SIRT7 peptide (K272ac/K275ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, NAD-dependent protein deacetylase sirtuin-7 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0R
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated POLR2A peptide (K775ac/K778ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DNA-directed RNA polymerase II subunit RPB1 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0O
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated ATRX peptide (K1030ac/K1033ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcriptional regulator ATRX | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0P
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated E2F1 peptide (K117ac/K120ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcription factor E2F1 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, Sorrell, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
3HP5
 
 | | Crystal Structure of Human p38alpha complexed with a pyrimidopyridazinone compound | | Descriptor: | 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, 5-(2,6-dichlorophenyl)-2-[(2,4-difluorophenyl)sulfanyl]-6H-pyrimido[1,6-b]pyridazin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Kurumbail, R.G. | | Deposit date: | 2009-06-03 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of N-substituted pyridinones as potent and selective inhibitors of p38 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6G0Q
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Erythroid transcription factor | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
3I3T
 
 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3HP2
 
 | | Crystal Structure of Human p38alpha complexed with a pyridinone compound | | Descriptor: | 1-benzyl-4-(benzyloxy)-3-bromopyridin-2(1H)-one, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Kurumbail, R.G. | | Deposit date: | 2009-06-03 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of N-substituted pyridinones as potent and selective inhibitors of p38 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8C3P
 
 | |
8C4W
 
 | | Crystal structure of rat autotaxin and compound MEY-002 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-[1-(phenylmethyl)indol-3-yl]chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C3O
 
 | | Crystal structure of autotaxin gamma and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
