6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NOU
 
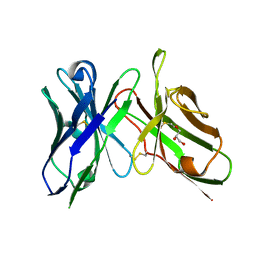 | | An scFv derived from ixekizumab | | Descriptor: | GLYCEROL, scFv derived from ixekizumab | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
6NOV
 
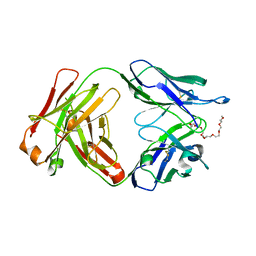 | | A Fab derived from ixekizumab | | Descriptor: | DODECAETHYLENE GLYCOL, Fab Heavy Chain, Fab Light Chain | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Schmitt, M, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
7M1S
 
 | |
4YCI
 
 | | non-latent pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, ... | | Authors: | Mi, L.Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YCG
 
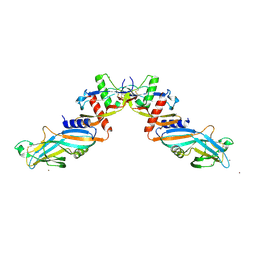 | | Pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, Bone Morphogenetic Protein 9 Prodomain, ... | | Authors: | Mi, L.-Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AC3
 
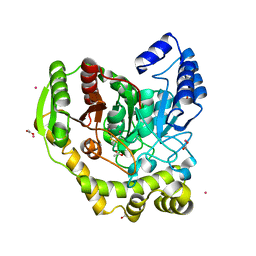 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
7S0N
 
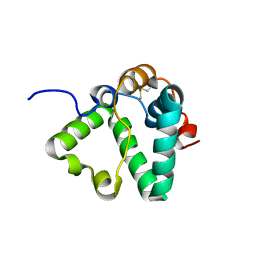 | | Structure of MS3494 from Mycobacterium Smegmatis determined by Solution NMR | | Descriptor: | Secreted protein | | Authors: | Kent, J.E, Tian, Y, Shin, K, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of MS3494 from Mycobacterium Smegmatis
To Be Published
|
|
7TDQ
 
 | |
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2LK9
 
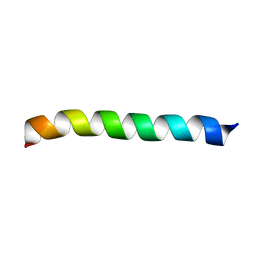 | | Structure of BST-2/Tetherin Transmembrane Domain | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Skasko, M, Wang, Y, Tian, Y, Tokarev, A, Munguia, J, Ruiz, A, Stephens, E, Opella, S, Guatelli, J. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions.
J.Biol.Chem., 287, 2012
|
|
6K9L
 
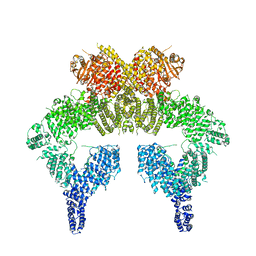 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
6IGM
 
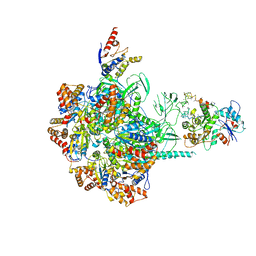 | | Cryo-EM Structure of Human SRCAP Complex | | Descriptor: | Actin-related protein 6, Helicase SRCAP, RuvB-like 1, ... | | Authors: | Feng, Y, Tian, Y, Wu, Z, Xu, Y. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of human SRCAP complex.
Cell Res., 28, 2018
|
|
2MCW
 
 | | Solid-state NMR structure of piscidin 3 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2N28
 
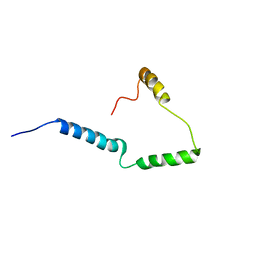 | | Solid-state NMR structure of Vpu | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2M67
 
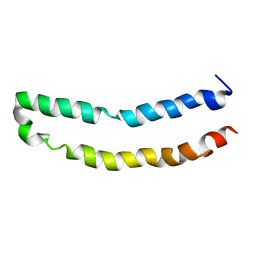 | | Full-length mercury transporter protein MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Lu, G.J, Tian, Y, Vora, N, Marassi, F.M, Opella, S.J. | | Deposit date: | 2013-03-27 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Structure of the Mercury Transporter MerF in Phospholipid Bilayers: A Large Conformational Rearrangement Results from N-Terminal Truncation.
J.Am.Chem.Soc., 135, 2013
|
|
2MCX
 
 | | Solid-state NMR structure of piscidin 3 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Wieczorek, W.E, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCU
 
 | | Solid-state NMR structure of piscidin 1 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2N29
 
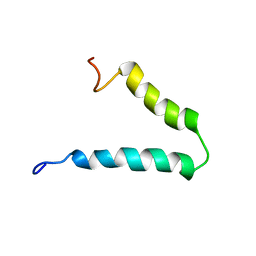 | | Solution-state NMR structure of Vpu cytoplasmic domain | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2MTS
 
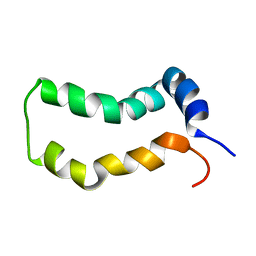 | | Three-Dimensional Structure and Interaction Studies of Hepatitis C Virus p7 in 1,2-Dihexanoyl-sn-glycero-3-phosphocholine by Solution Nuclear Magnetic Resonance | | Descriptor: | HEPATITIS C VIRUS P7 PROTEIN | | Authors: | Cook, G.A, Dawson, L.A, Tian, Y, Opella, S.J. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance.
Biochemistry, 52, 2013
|
|
6X7I
 
 | |
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
5YZ0
 
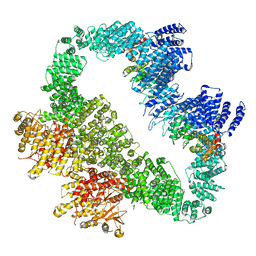 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
5Y3R
 
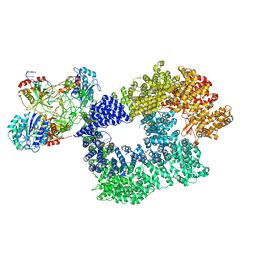 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | Descriptor: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | Deposit date: | 2017-07-29 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
