2C8D
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8F
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (NAD-bound state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8A
 
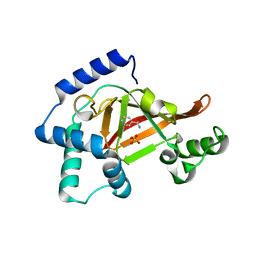 | | Structure of the wild-type C3bot1 Exoenzyme (Nicotinamide-bound state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8C
 
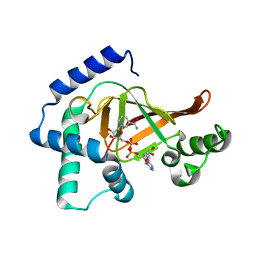 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
5K1N
 
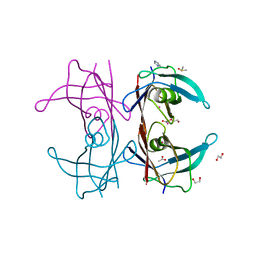 | | Human TTR altered by a rhenium tris-carbonyl Pyta-C12 derivative | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Ciccone, L, Shepard, W. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Human TTR conformation altered by rhenium tris-carbonyl derivatives.
J.Struct.Biol., 195, 2016
|
|
5K1J
 
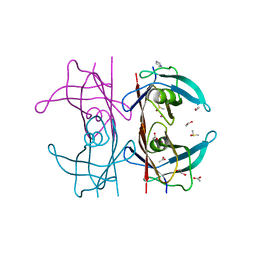 | | Human TTR altered by a rhenium tris-carbonyl Pyta-C8 derivative | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Stura, E.A, Ciccone, L, Shepard, W. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Human TTR conformation altered by rhenium tris-carbonyl derivatives.
J.Struct.Biol., 195, 2016
|
|
5I0L
 
 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
3F5V
 
 | | C2 Crystal form of mite allergen DER P 1 | | Descriptor: | CALCIUM ION, Der p 1 allergen, HEXAETHYLENE GLYCOL | | Authors: | Stura, E.A, Minor, W, Chruszcz, M, Saint Remy, J.M. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding.
J.Mol.Biol., 386, 2009
|
|
3KZ7
 
 | |
3FEV
 
 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 1 from MT1. | | Descriptor: | Fusion of Muscarinic toxin 1, Muscarinic m1-toxin1, SULFATE ION | | Authors: | Stura, E.A, Menez, R, Mourier, G, Fruchart-Gaillard, C, Menez, A, Servant, D. | | Deposit date: | 2008-12-01 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
5MG9
 
 | | Putative Ancestral Mamba toxin 1 (AncTx1-W28R/I38S) | | Descriptor: | 1,2-ETHANEDIOL, AncTx1-W28R/I38S, S-1,2-PROPANEDIOL, ... | | Authors: | Stura, E.A, Tepshi, L, Blanchet, G, Mourier, G, Servent, D. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Ancestral protein resurrection and engineering opportunities of the mamba aminergic toxins.
Sci Rep, 7, 2017
|
|
5M4V
 
 | | X-ray structure of the mambaquaretin-1, a selective antagonist of the vasopressin type 2 receptor | | Descriptor: | CHLORIDE ION, Mambaquaretin-1, S-1,2-PROPANEDIOL | | Authors: | Stura, E.A, Vera, L, Ciolek, J, Mourier, G, Gilles, N. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3NEQ
 
 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 3 from MT1 | | Descriptor: | SULFATE ION, Three-finger muscarinic toxin 7 | | Authors: | Stura, E.A, Servent, D, Menez, R, Mournier, G, Menez, A, Fruchart-Gaillard, C. | | Deposit date: | 2010-06-09 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
5DU1
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P21 space group. | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
5DZ5
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P41212 space group | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
4H3X
 
 | | Crystal structure of an MMP broad spectrum hydroxamate based inhibitor CC27 in complex with the MMP-9 catalytic domain | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4H76
 
 | | Crystal structure of the catalytic domain of Human MMP12 in complex with a broad spectrum hydroxamate inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4H84
 
 | | Crystal structure of the catalytic domain of Human MMP12 in complex with a selective carboxylate based inhibitor. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Antoni, C, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-21 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
4EFS
 
 | | Human MMP12 in complex with L-glutamate motif inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
3TS4
 
 | | Human MMP12 in complex with L-glutamate motif inhibitor | | Descriptor: | CALCIUM ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L. | | Deposit date: | 2011-09-12 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
3TVC
 
 | | Human MMP13 in complex with L-glutamate motif inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L, Cassar-Lajeunesse, E. | | Deposit date: | 2011-09-20 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
3TT4
 
 | | Human MMP8 in complex with L-glutamate motif inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L. | | Deposit date: | 2011-09-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
3TSK
 
 | | Human MMP12 in complex with L-glutamate motif inhibitor | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N~2~-{3-[4-(4-phenylthiophen-2-yl)phenyl]propanoyl}-L-glutaminyl-L-alpha-glutamine, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
