3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1Z5P
 
 | | Crystal structure of MTA/AdoHcy nucleosidase with a ligand-free purine binding site | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5O
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
2OLC
 
 | | Crystal structure of 5-methylthioribose kinase in complex with ADP-2Ho | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ADENOSINE-5'-DIPHOSPHATE, HOLMIUM ATOM, ... | | Authors: | Ku, S.Y, Smith, G.D, Howell, P.L. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-2Ho as a phasing tool for nucleotide-containing proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3DF9
 
 | |
6I3U
 
 | |
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
5M5K
 
 | | S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine and cordycepin | | Descriptor: | 3'-DEOXYADENOSINE, ACETATE ION, ADENOSINE, ... | | Authors: | Manszewski, T, Mueller-Dieckamann, J, Jaskolski, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
5M65
 
 | |
5M66
 
 | |
5M67
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenine and 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, ACETATE ION, ADENINE, ... | | Authors: | Manszewski, T, Jaskolski, M. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
1MPJ
 
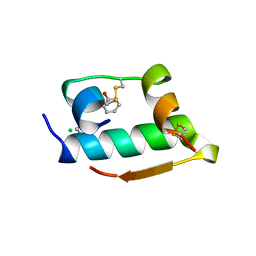 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1ZNI
 
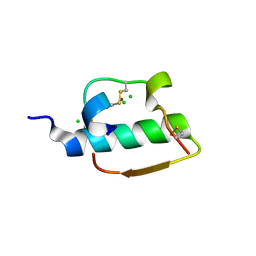 | | INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, ZINC ION | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Dodson, G.G, Dodson, E.J, Xiao, B, Bentley, G.A. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of insulin in 4-zinc insulin.
Nature, 261, 1976
|
|
1RWE
 
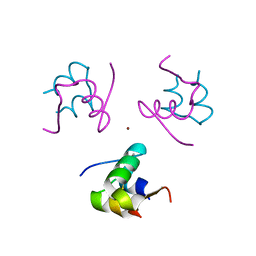 | | Enhancing the activity of insulin at receptor edge: crystal structure and photo-cross-linking of A8 analogues | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z, Xu, B, Chu, Y.C, Li, B, Nakagawa, S.H, Qu, Y, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues.
Biochemistry, 43, 2004
|
|
1XDA
 
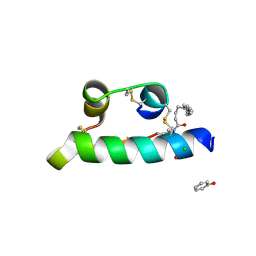 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, FATTY ACID ACYLATED INSULIN, MYRISTIC ACID, ... | | Authors: | Whittingham, J.L, Havelund, S, Jonassen, I. | | Deposit date: | 1996-12-18 | | Release date: | 1997-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a prolonged-acting insulin with albumin-binding properties.
Biochemistry, 36, 1997
|
|
7INS
 
 | | STRUCTURE OF PORCINE INSULIN COCRYSTALLIZED WITH CLUPEINE Z | | Descriptor: | GENERAL PROTAMINE CHAIN, INSULIN (CHAIN A), INSULIN (CHAIN B), ... | | Authors: | Balschmidt, P, Hansen, F.B, Dodson, E, Dodson, G, Korber, F. | | Deposit date: | 1991-09-03 | | Release date: | 1994-01-31 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of porcine insulin cocrystallized with clupeine Z.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
3MTH
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEH
 
 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEI
 
 | | CROSS-LINKED B28 ASP INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
2TCI
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | THIOCYANATE INSULIN, THIOCYANATE ION, ZINC ION | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1SF7
 
 | | BINDING OF TETRA-N-ACETYLCHITOTETRAOSE TO HEW LYSOZYME: A POWDER DIFFRACTION STUDY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Von Dreele, R.B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | POWDER DIFFRACTION | | Cite: | Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: a powder diffraction study.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SF4
 
 | |
1SFB
 
 | | BINDING OF PENTA-N-ACETYLCHITOPENTAOSE TO HEW LYSOZYME: A POWDER DIFFRACTION STUDY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Von Dreele, R.B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | POWDER DIFFRACTION | | Cite: | Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: a powder diffraction study.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SF6
 
 | | BINDING OF N,N',N"-TRIACETYLCHITOTRIOSE TO HEW LYSOZYME: A POWDER DIFFRACTION STUDY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Von Dreele, R.B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-13 | | Method: | POWDER DIFFRACTION | | Cite: | Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: a powder diffraction study.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
